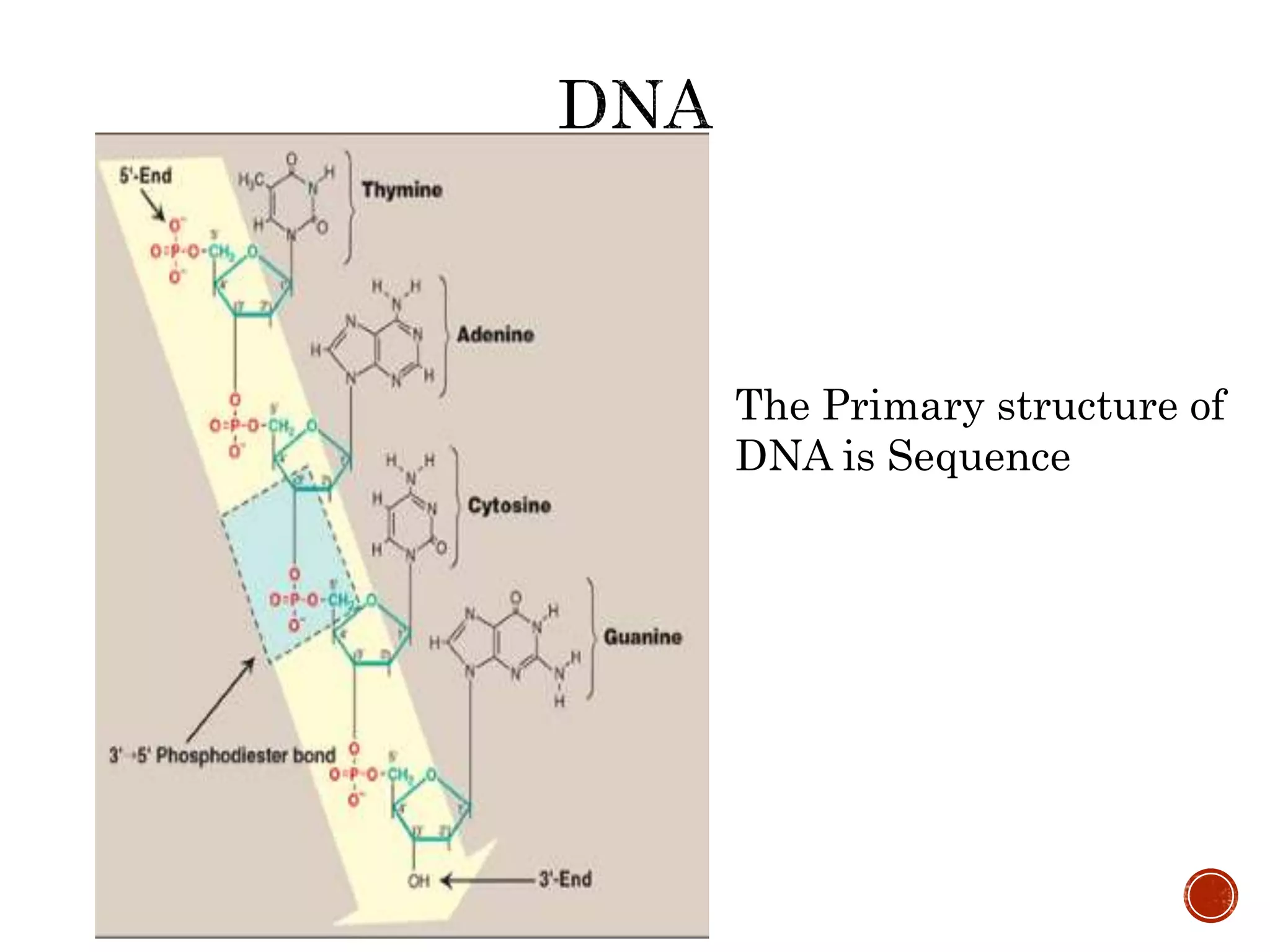
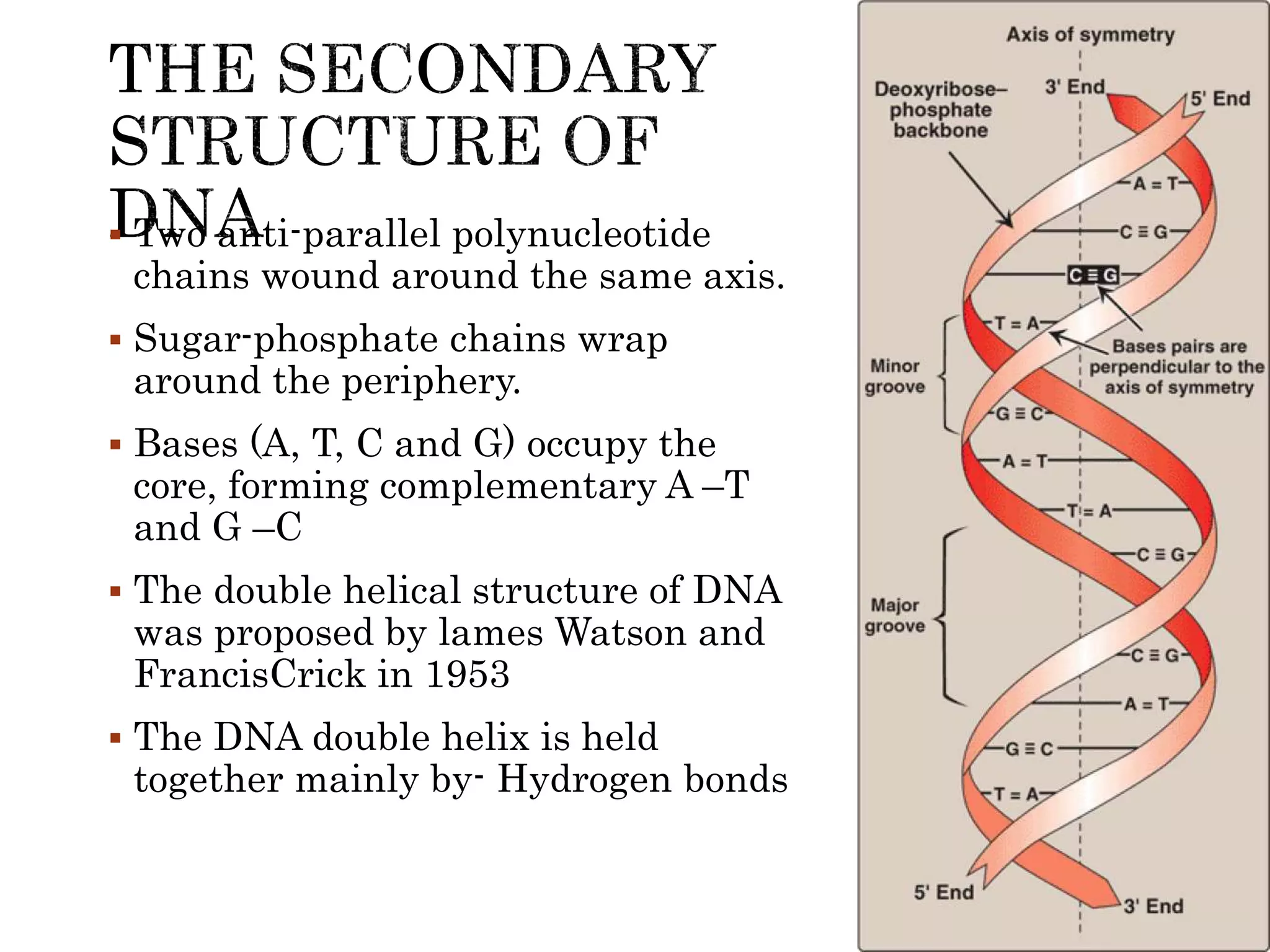
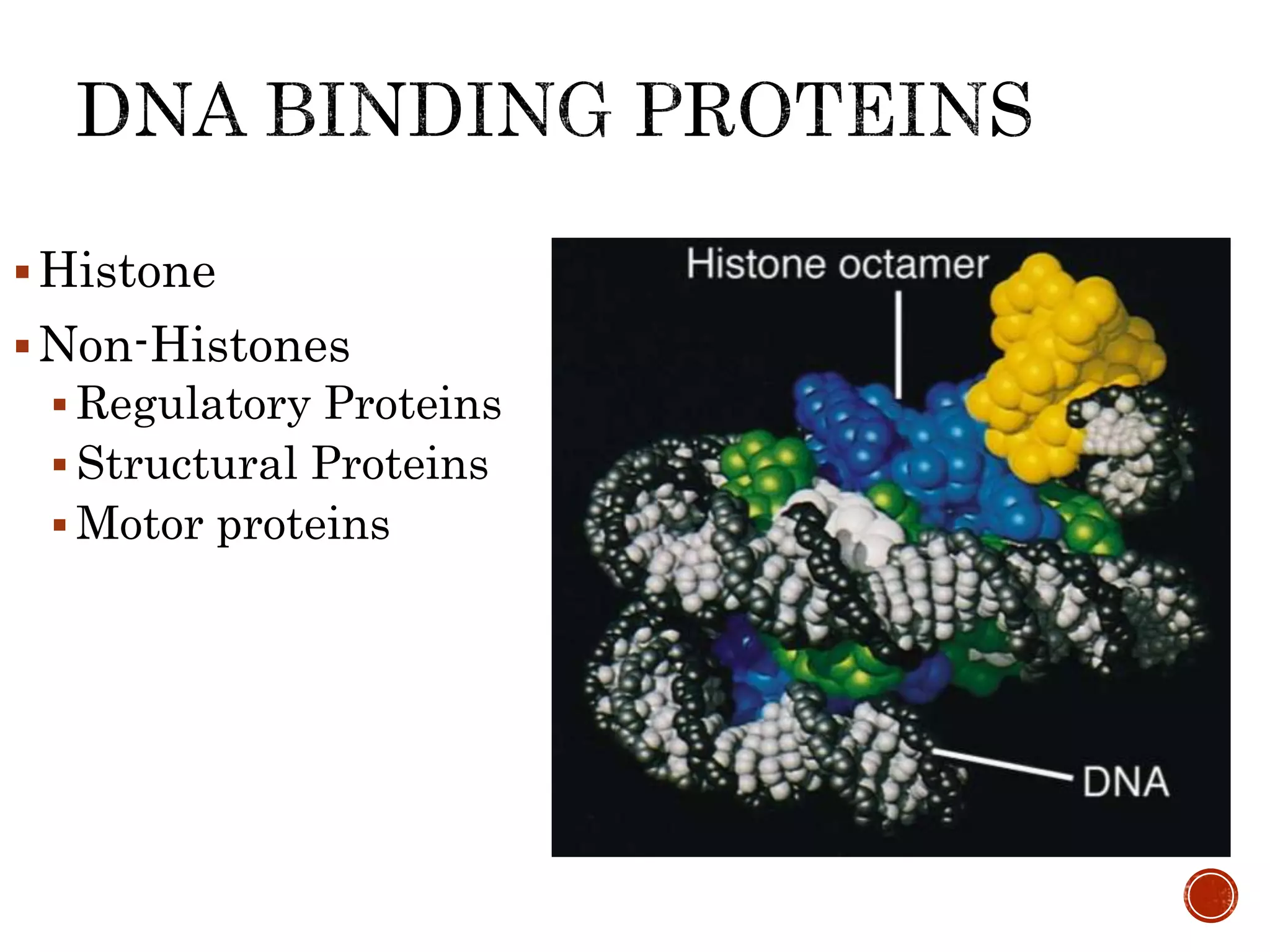
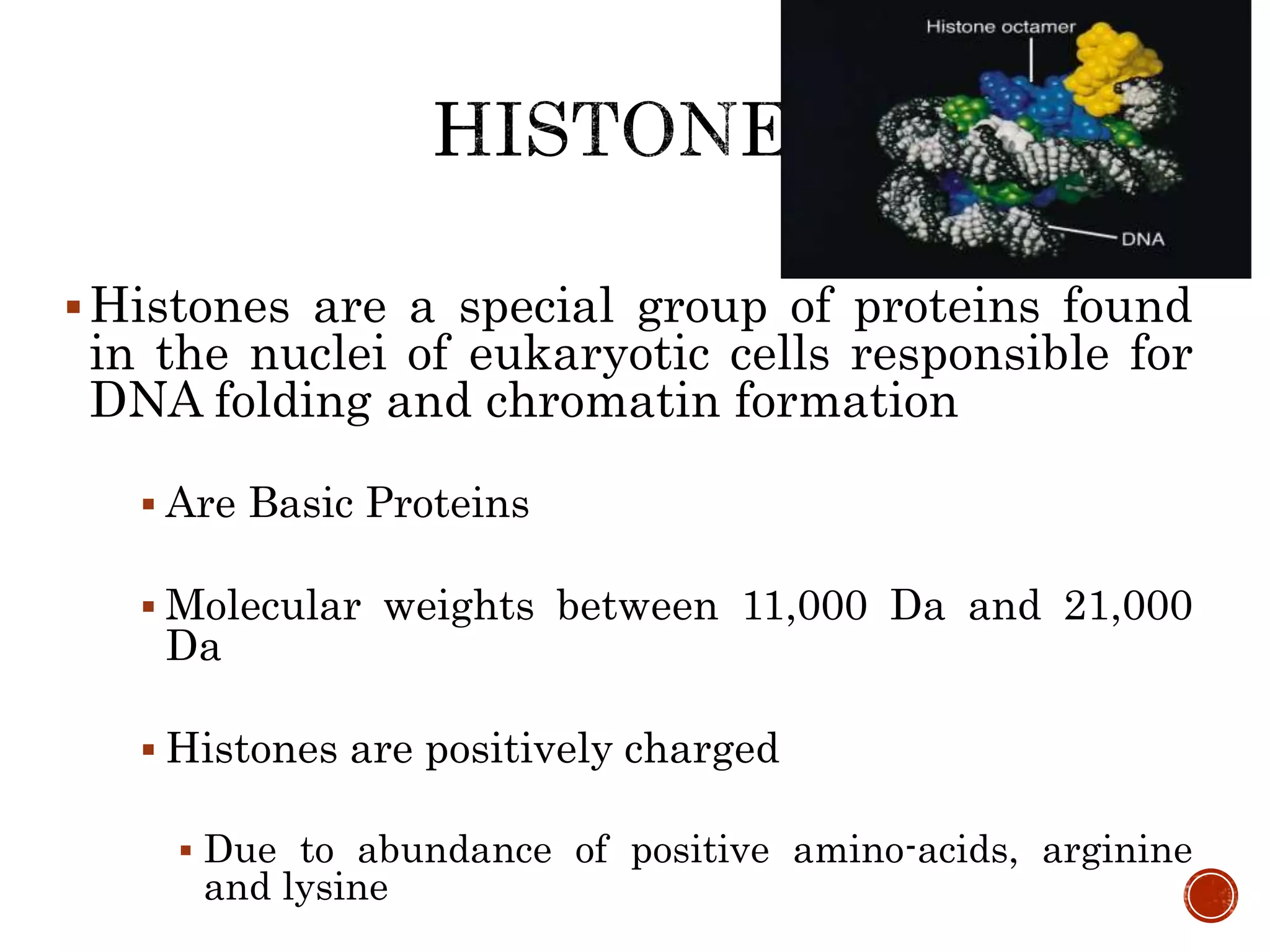
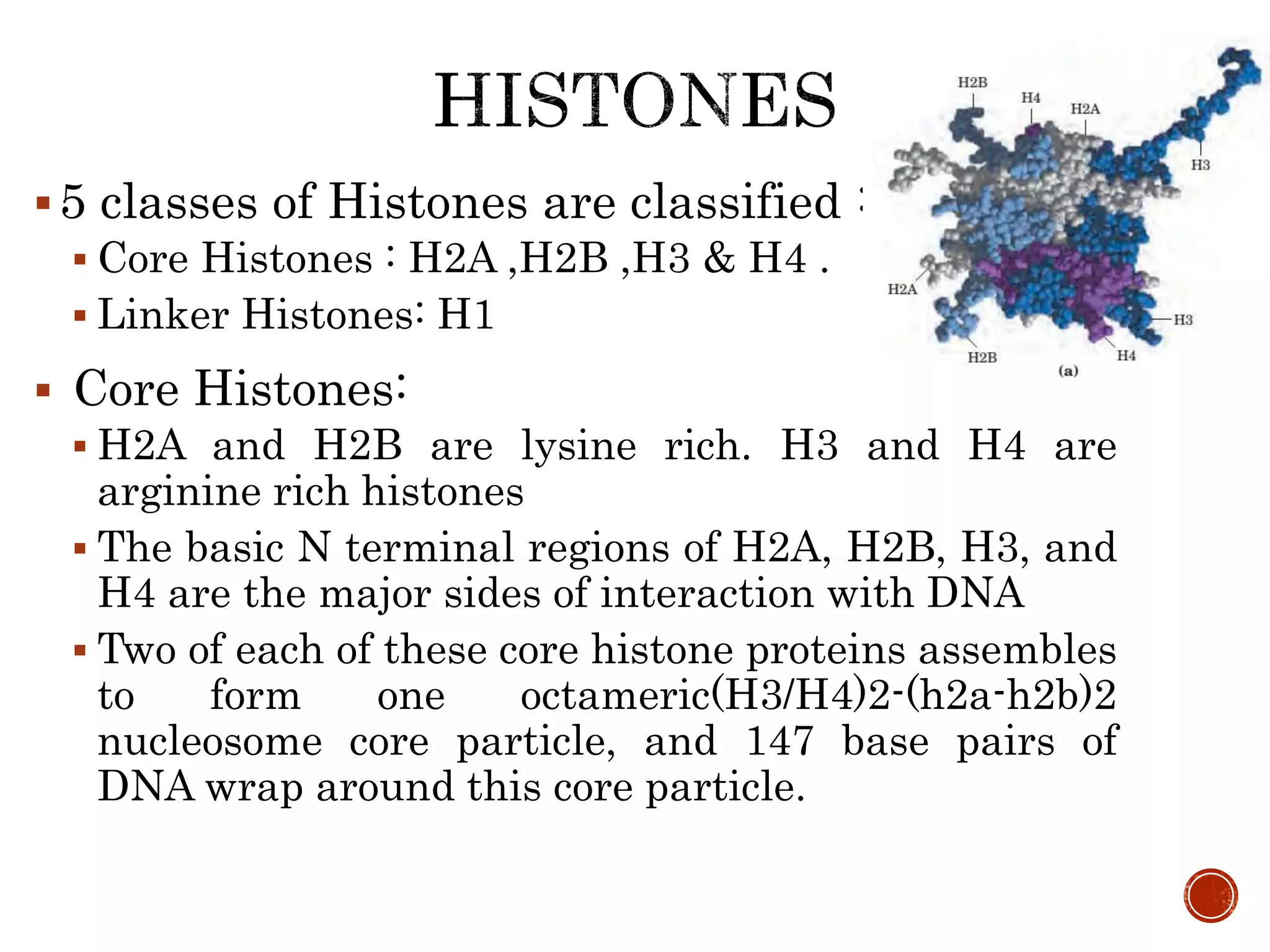
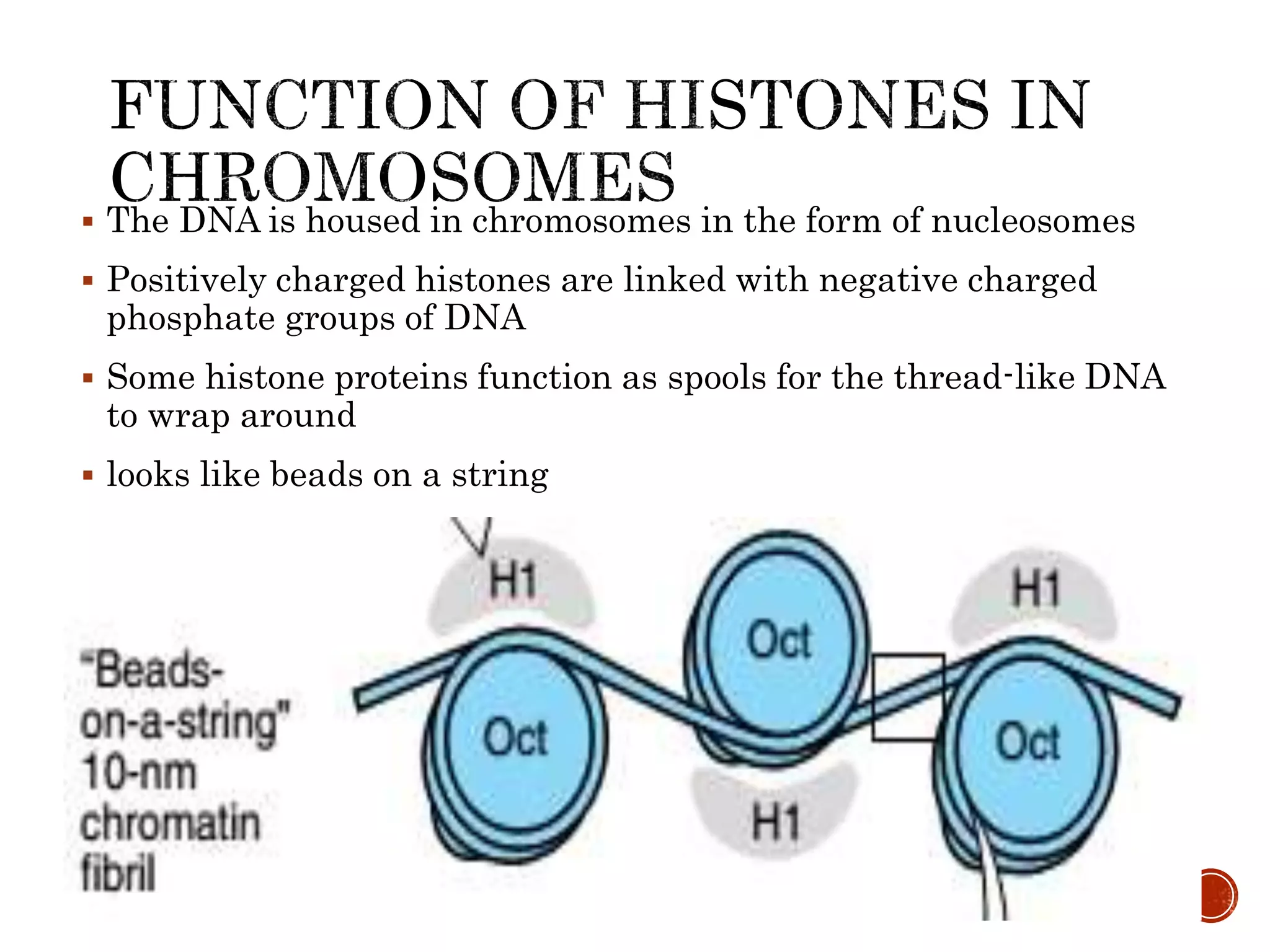
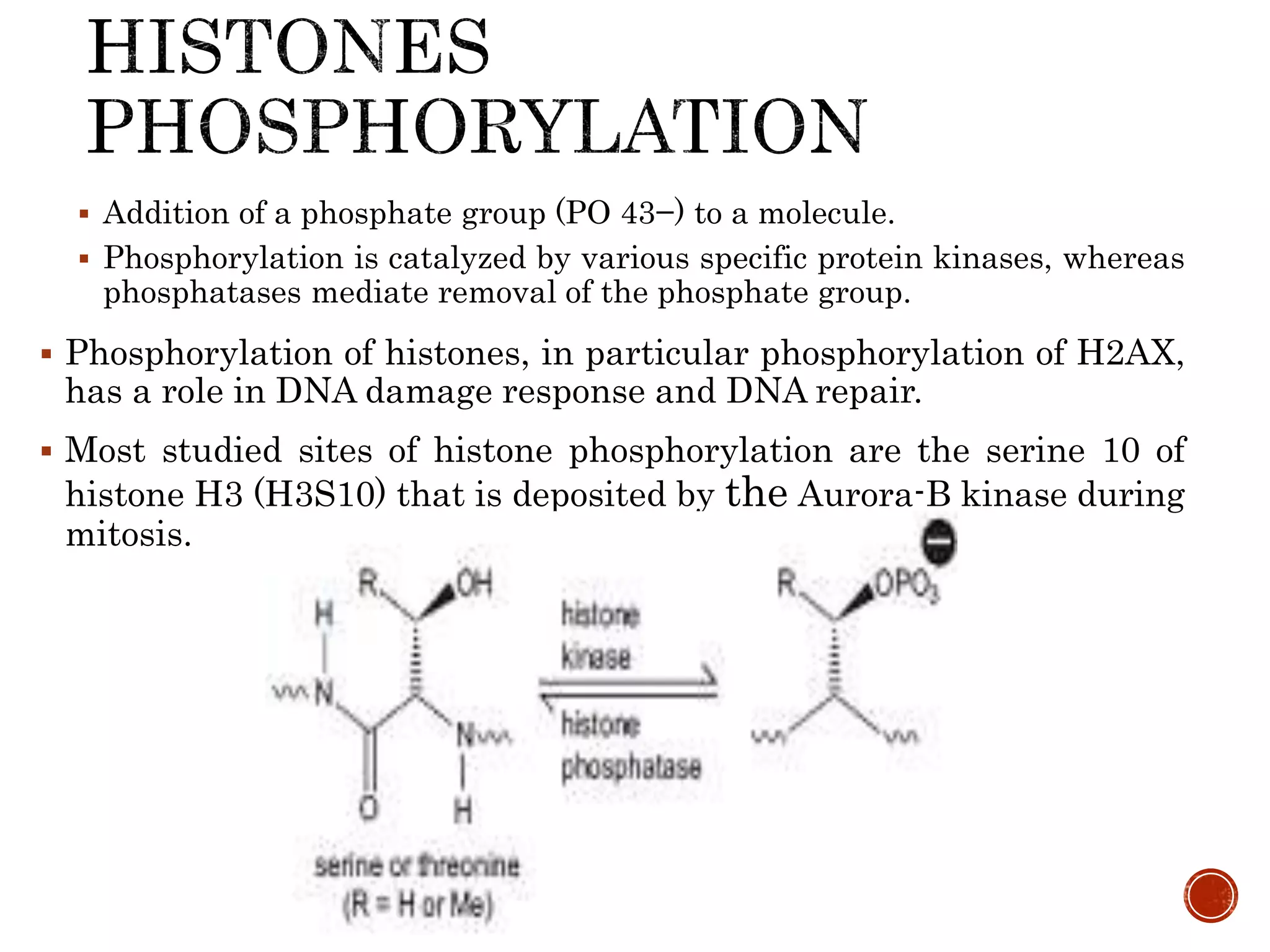
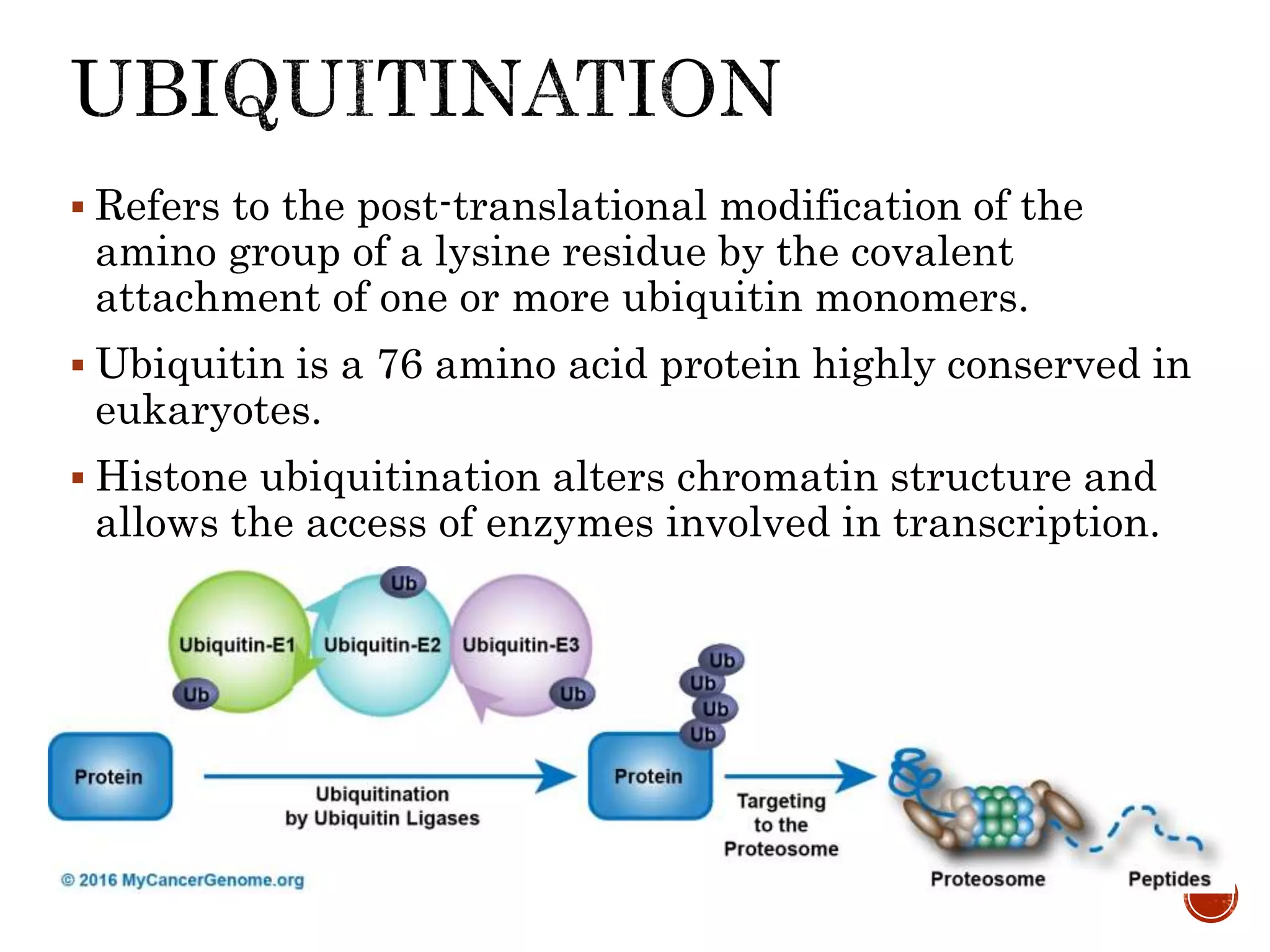
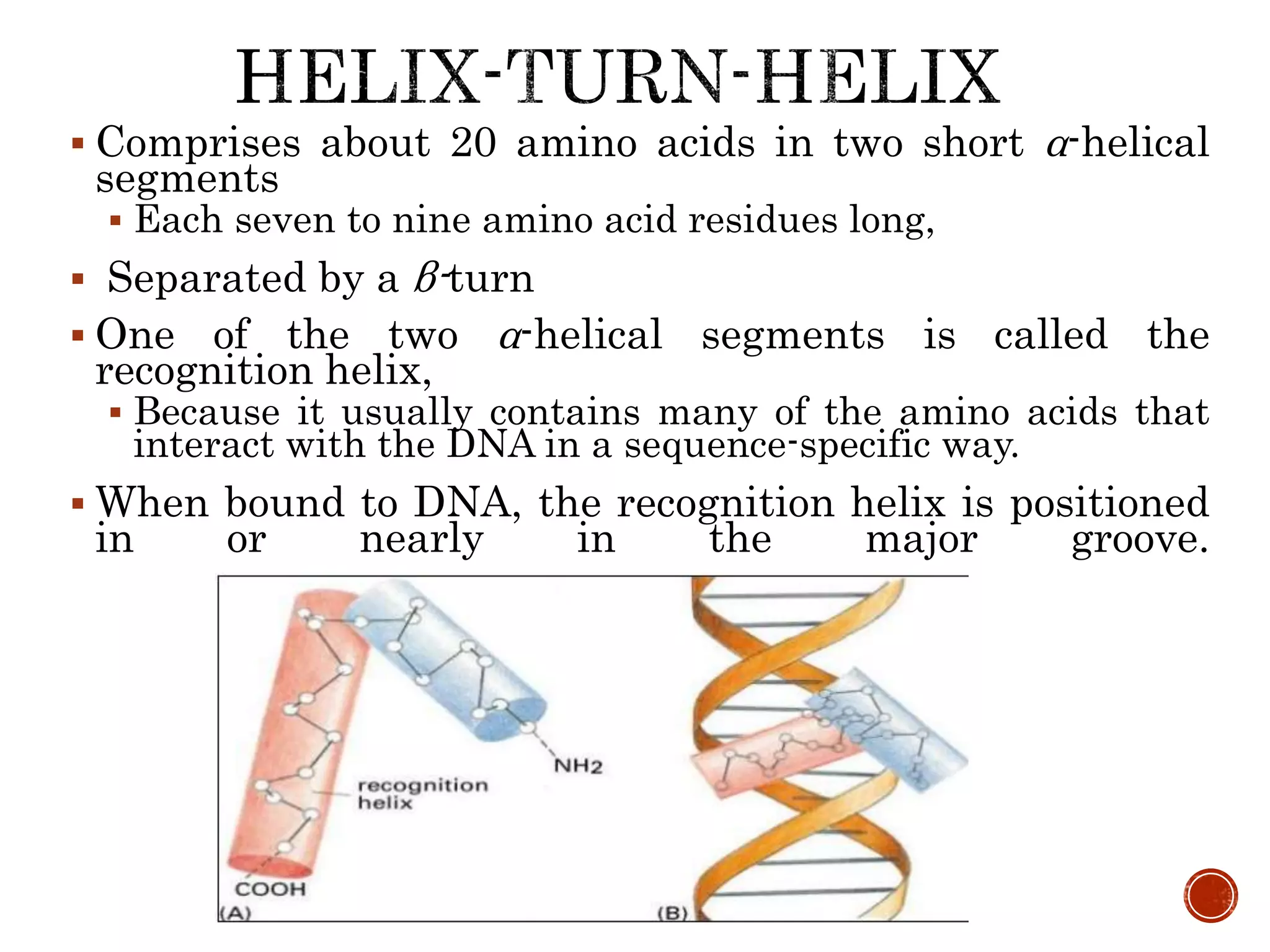
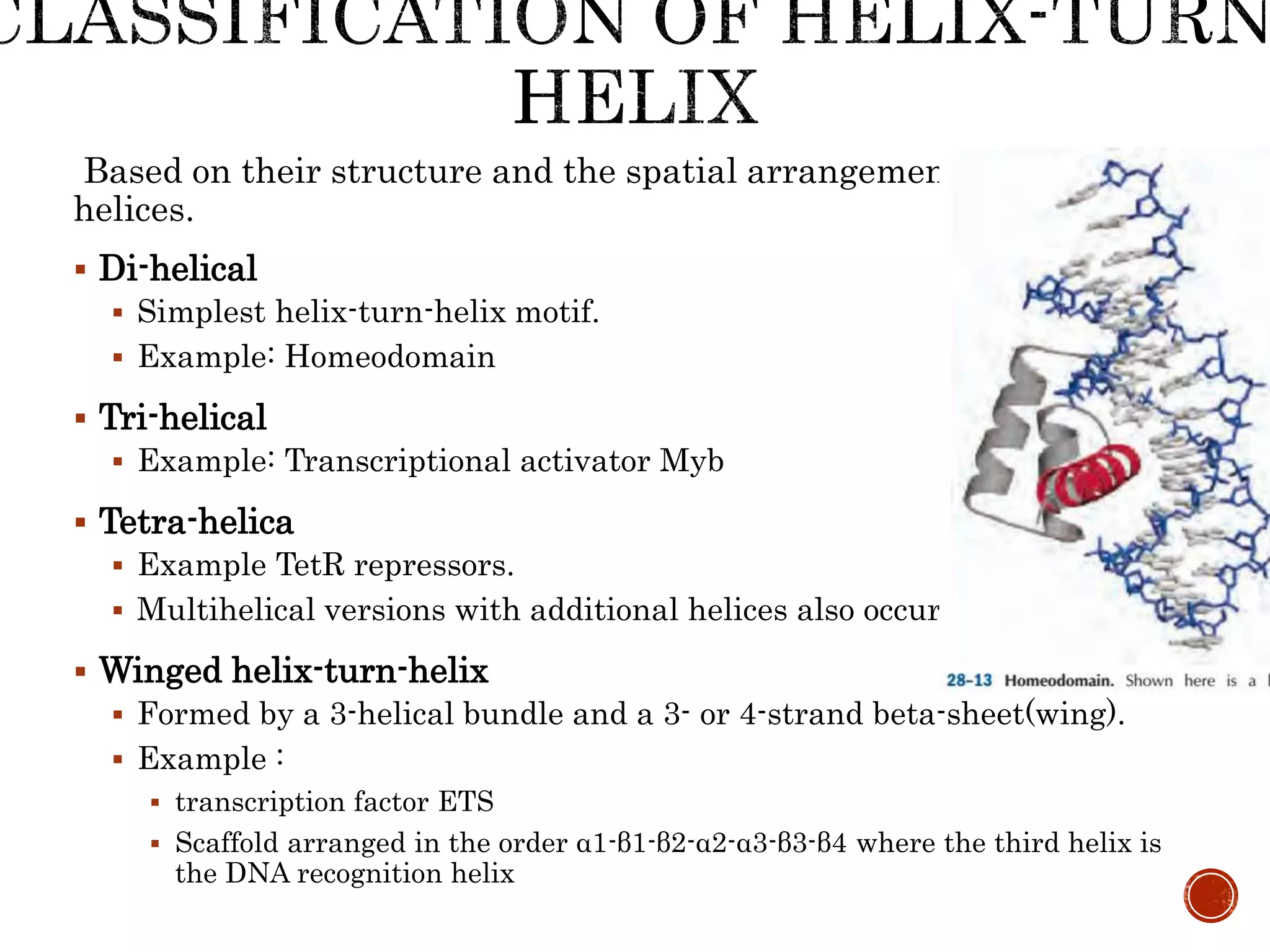
The document discusses DNA binding proteins. It describes how DNA is wrapped around histone proteins to form nucleosomes, which resemble "beads on a string". There are five main types of histone proteins - H1, H2A, H2B, H3, and H4. Histone proteins can be modified through processes like acetylation and methylation, which affect gene expression. Other non-histone proteins use motifs like zinc fingers and helix-turn-helix to bind DNA in a sequence-specific manner and regulate transcription.