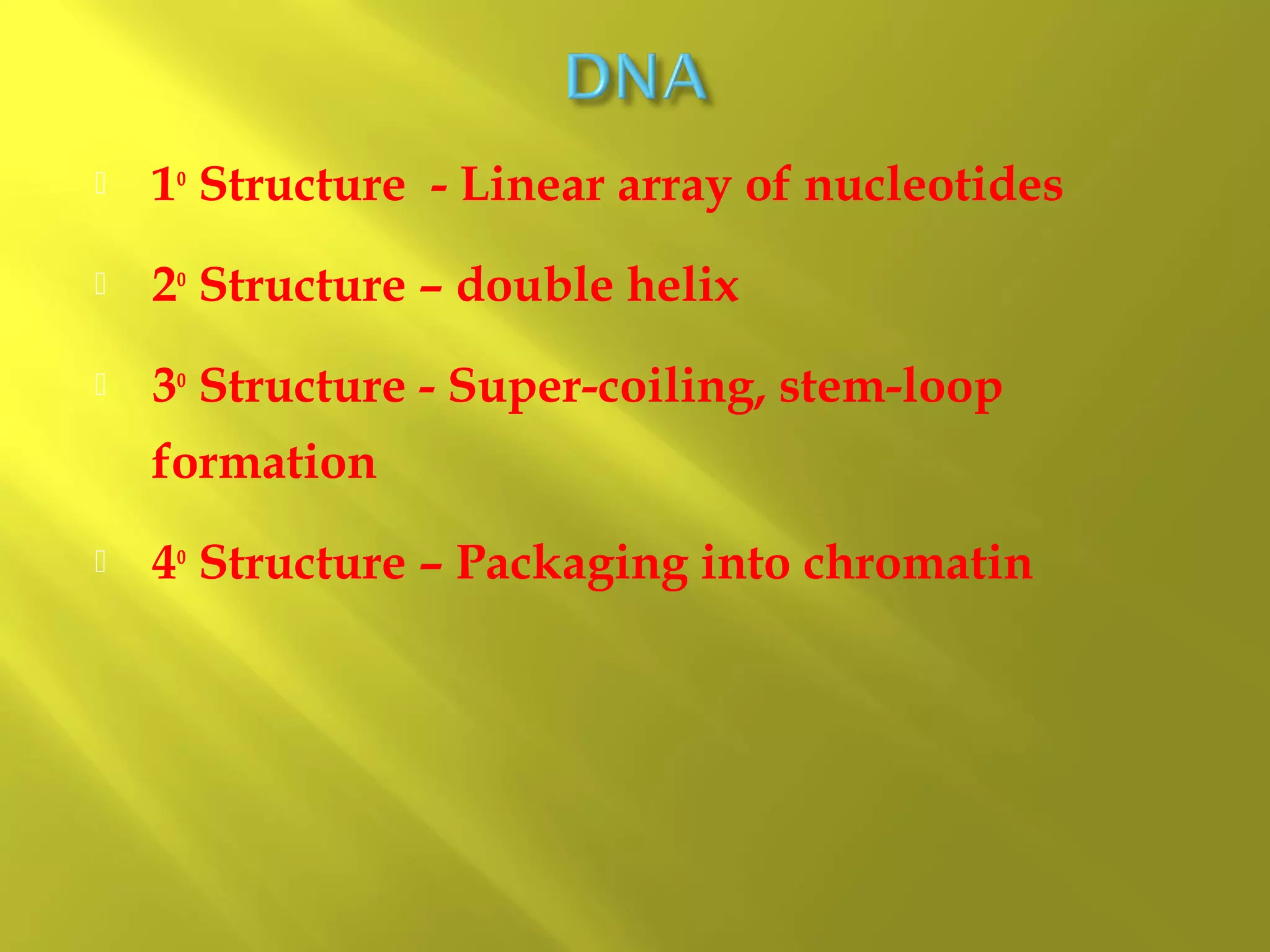
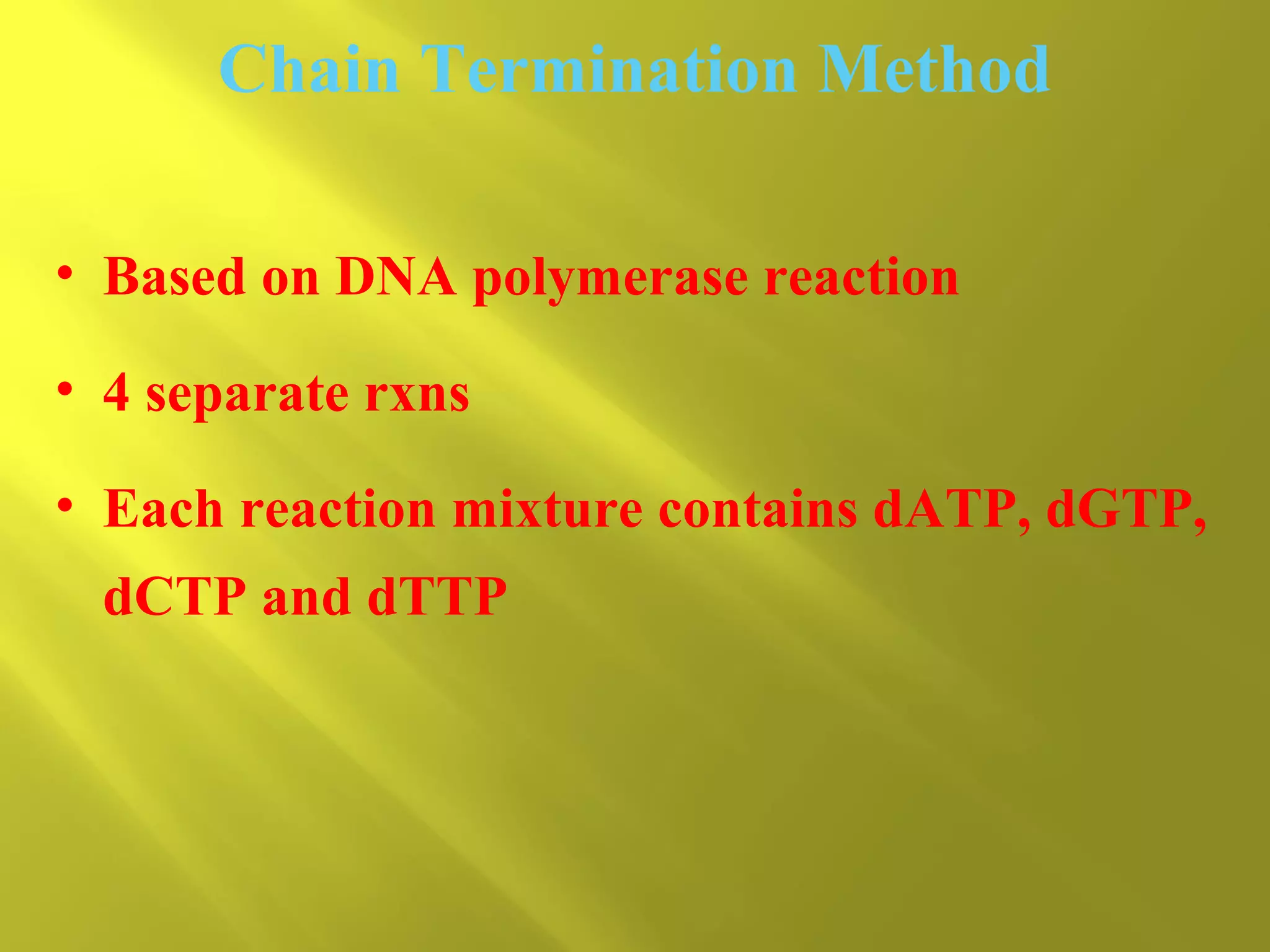
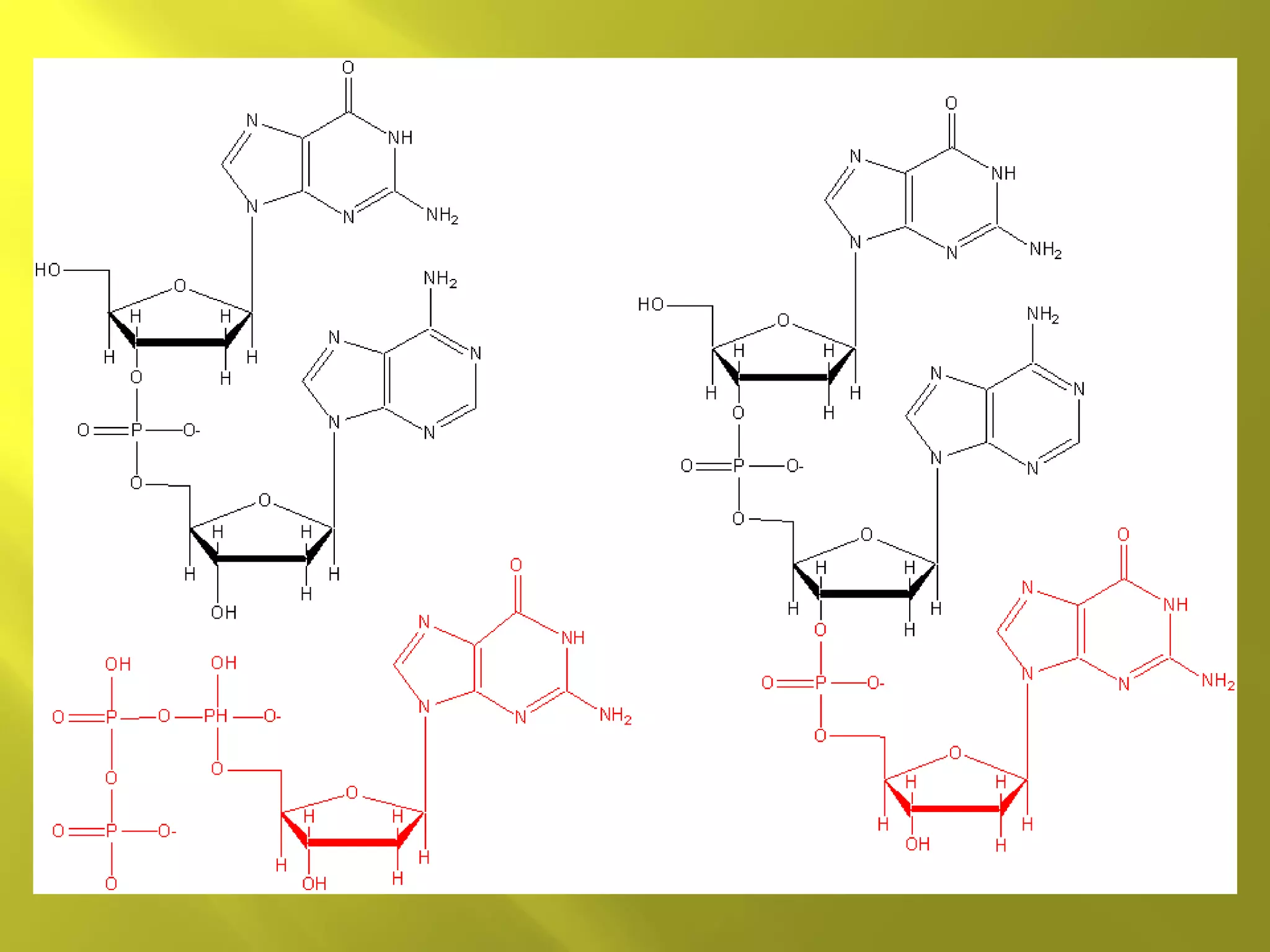
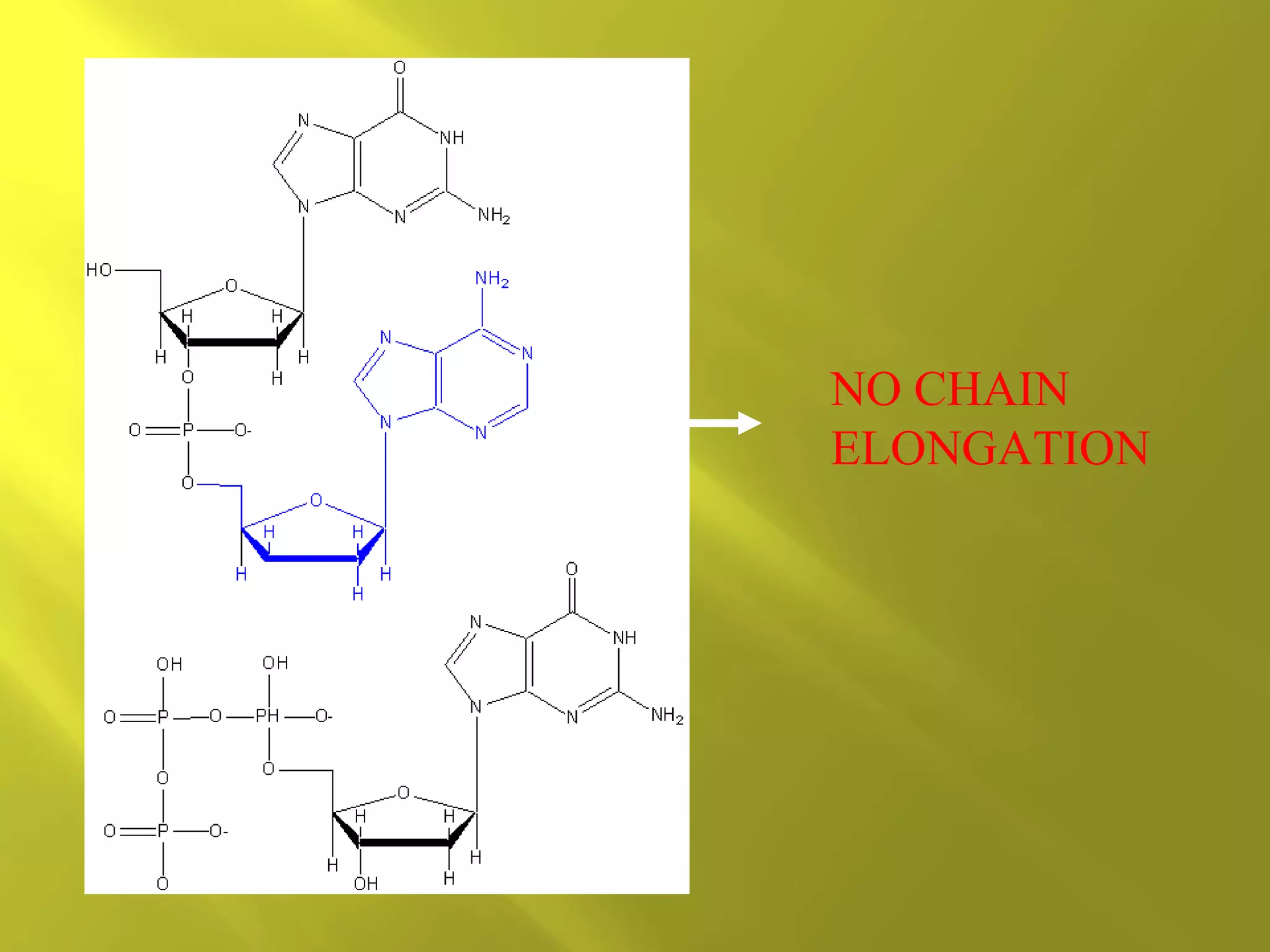
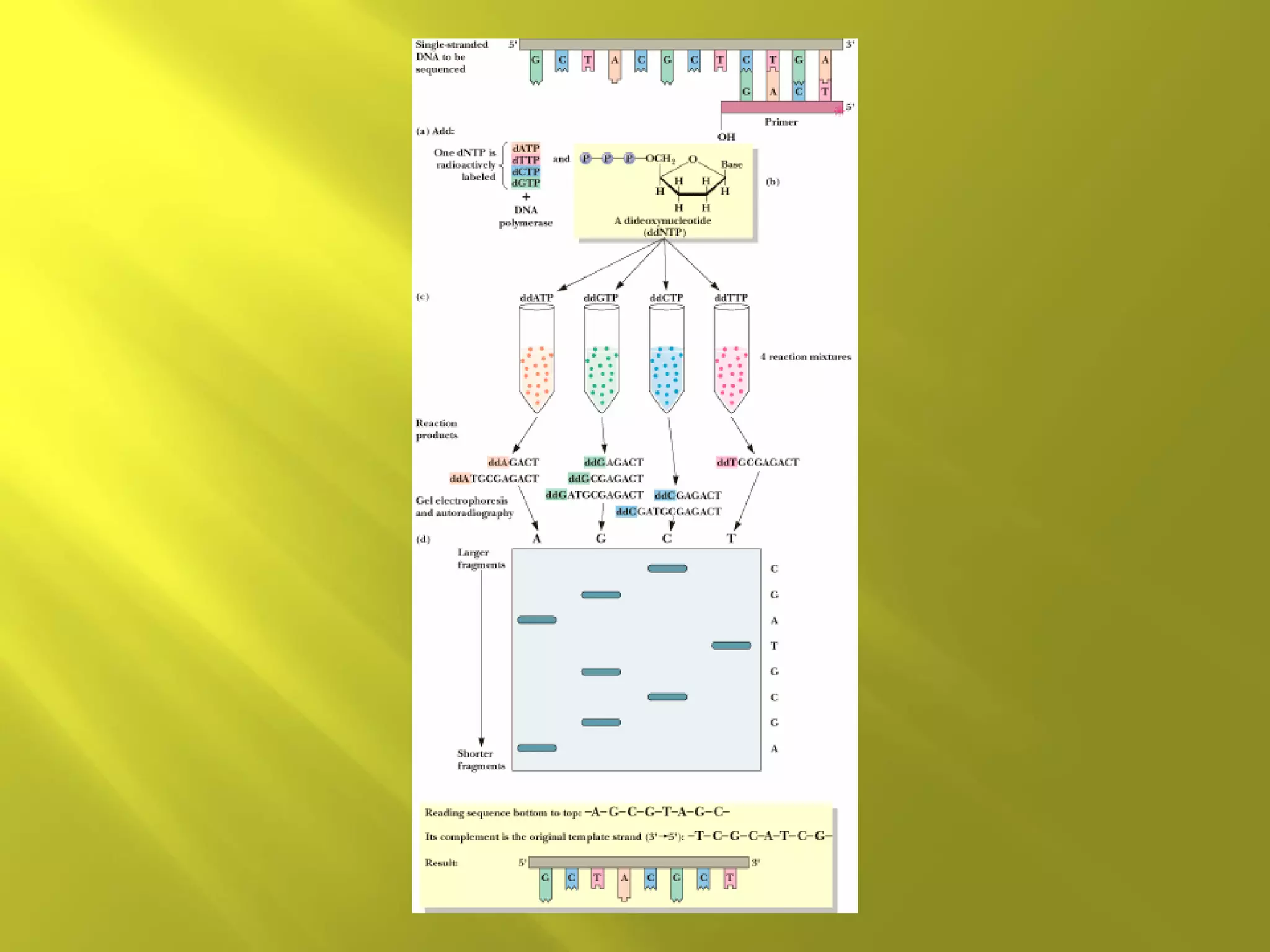
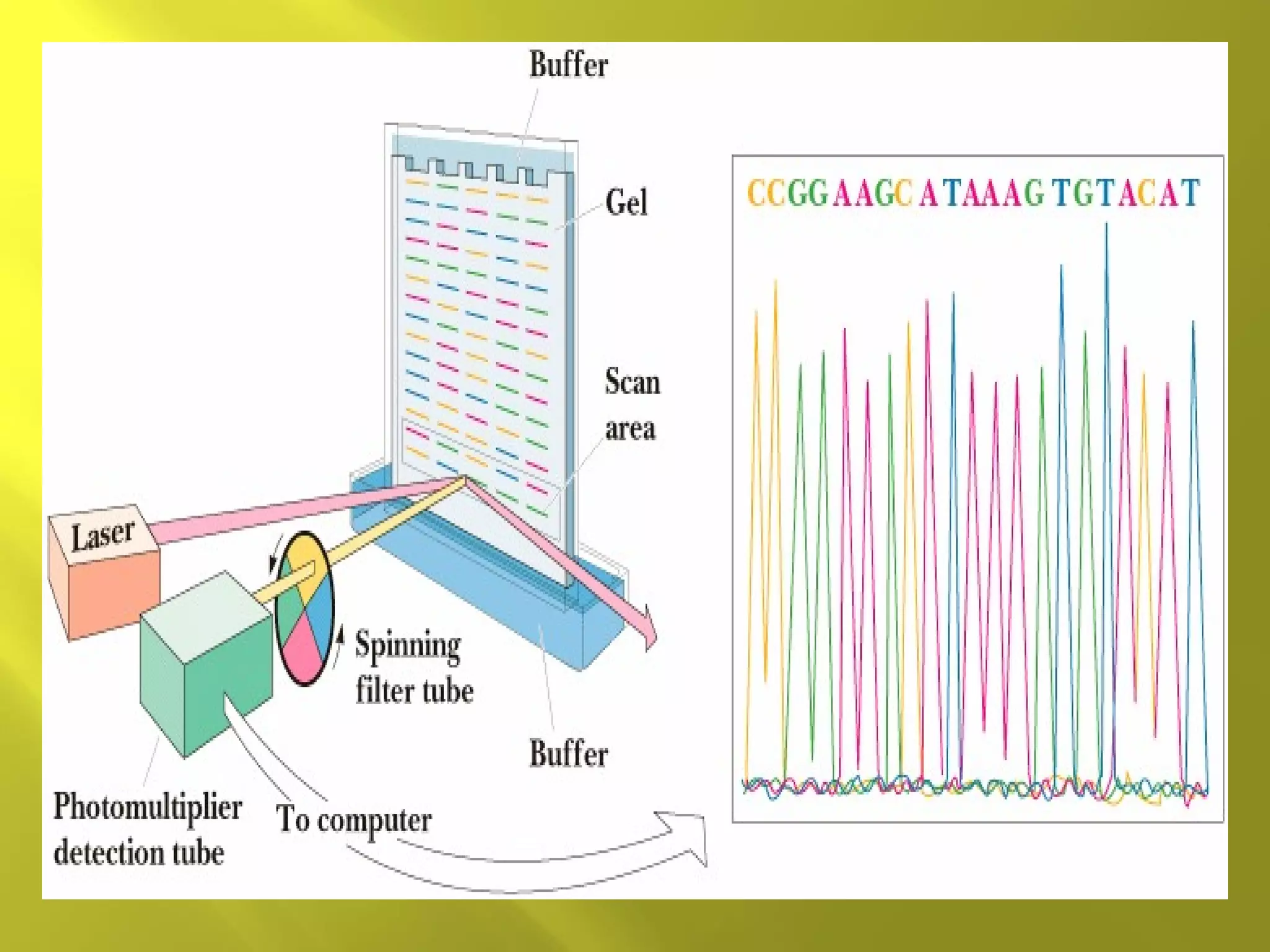
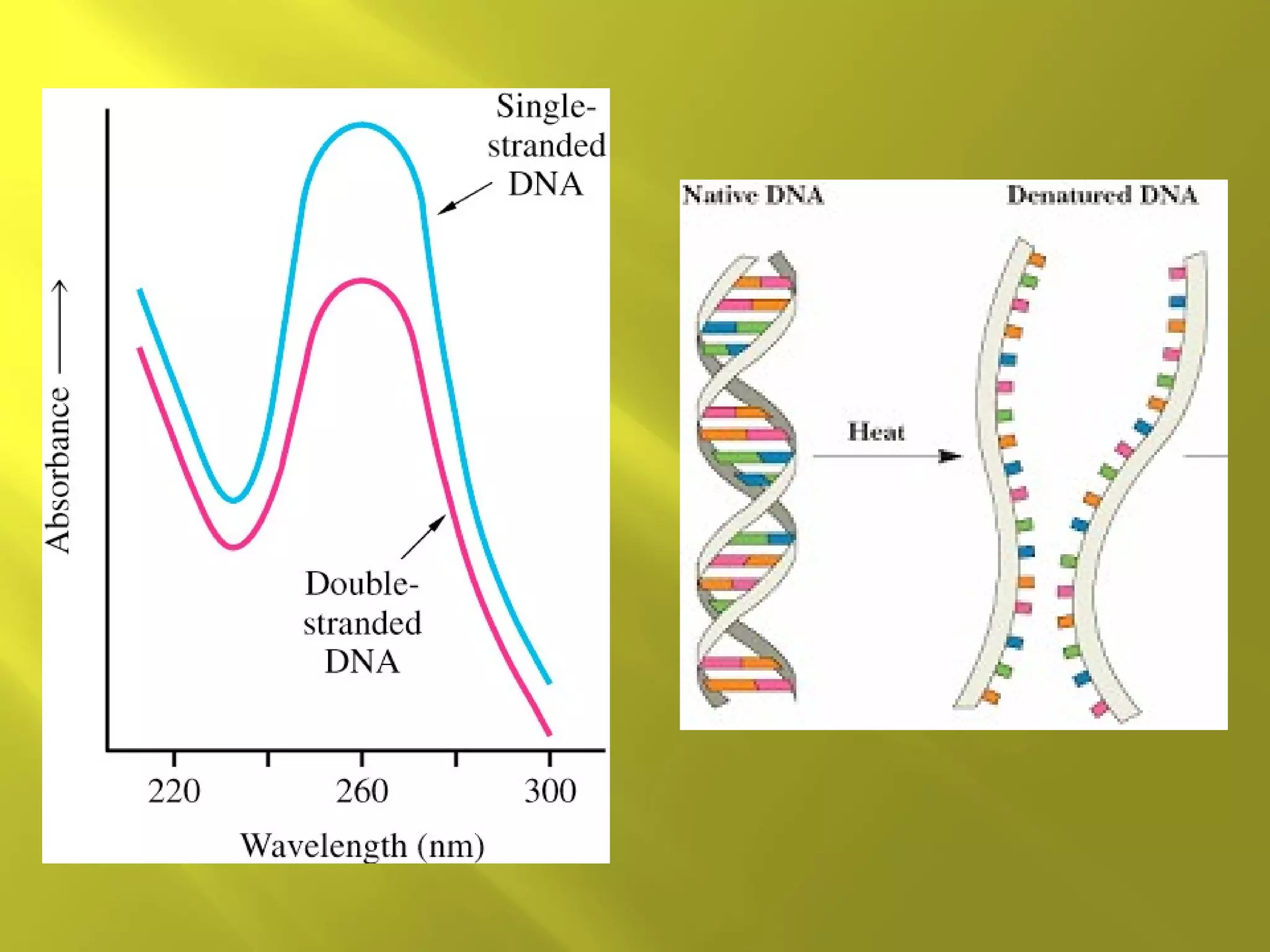
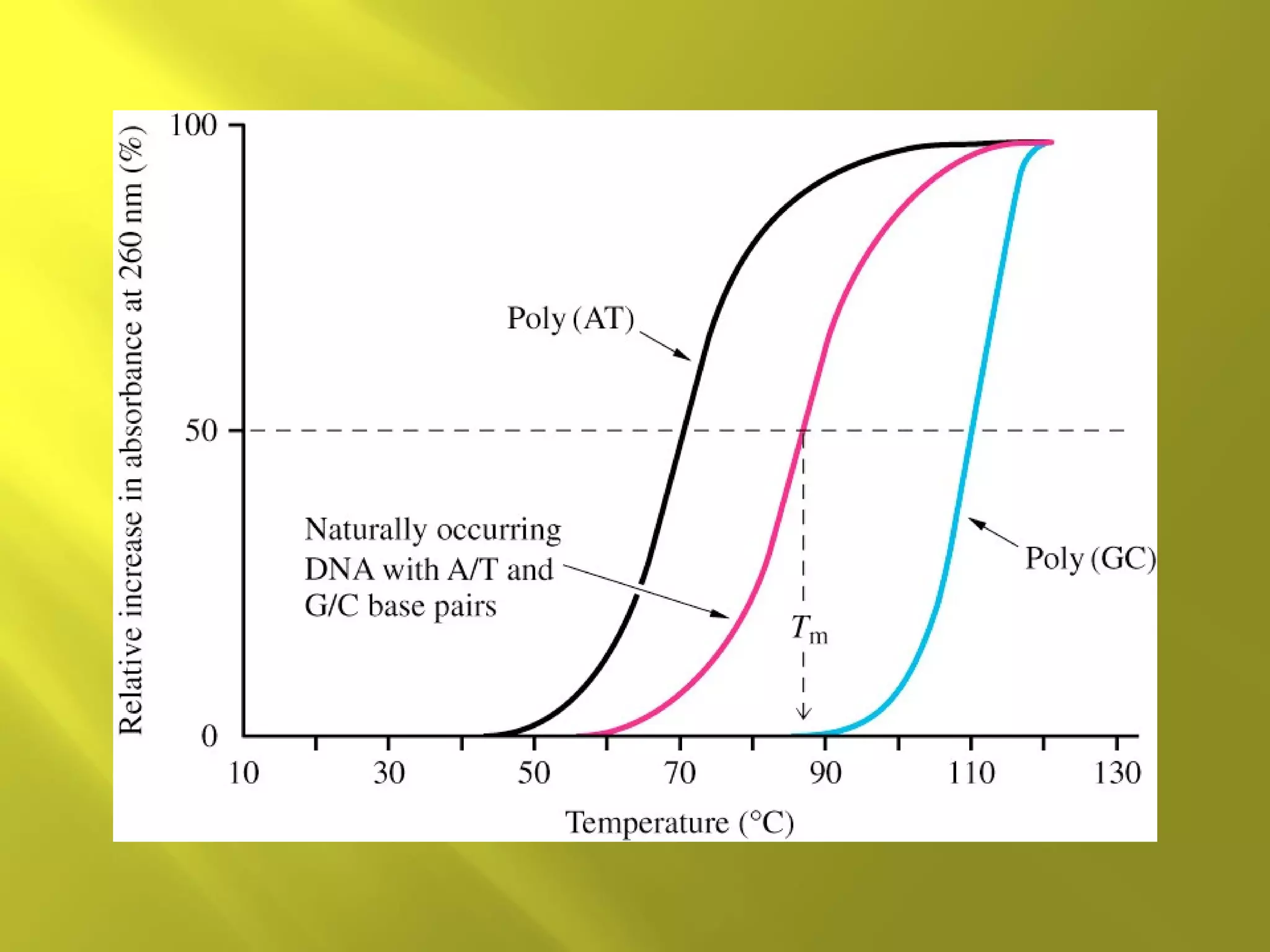
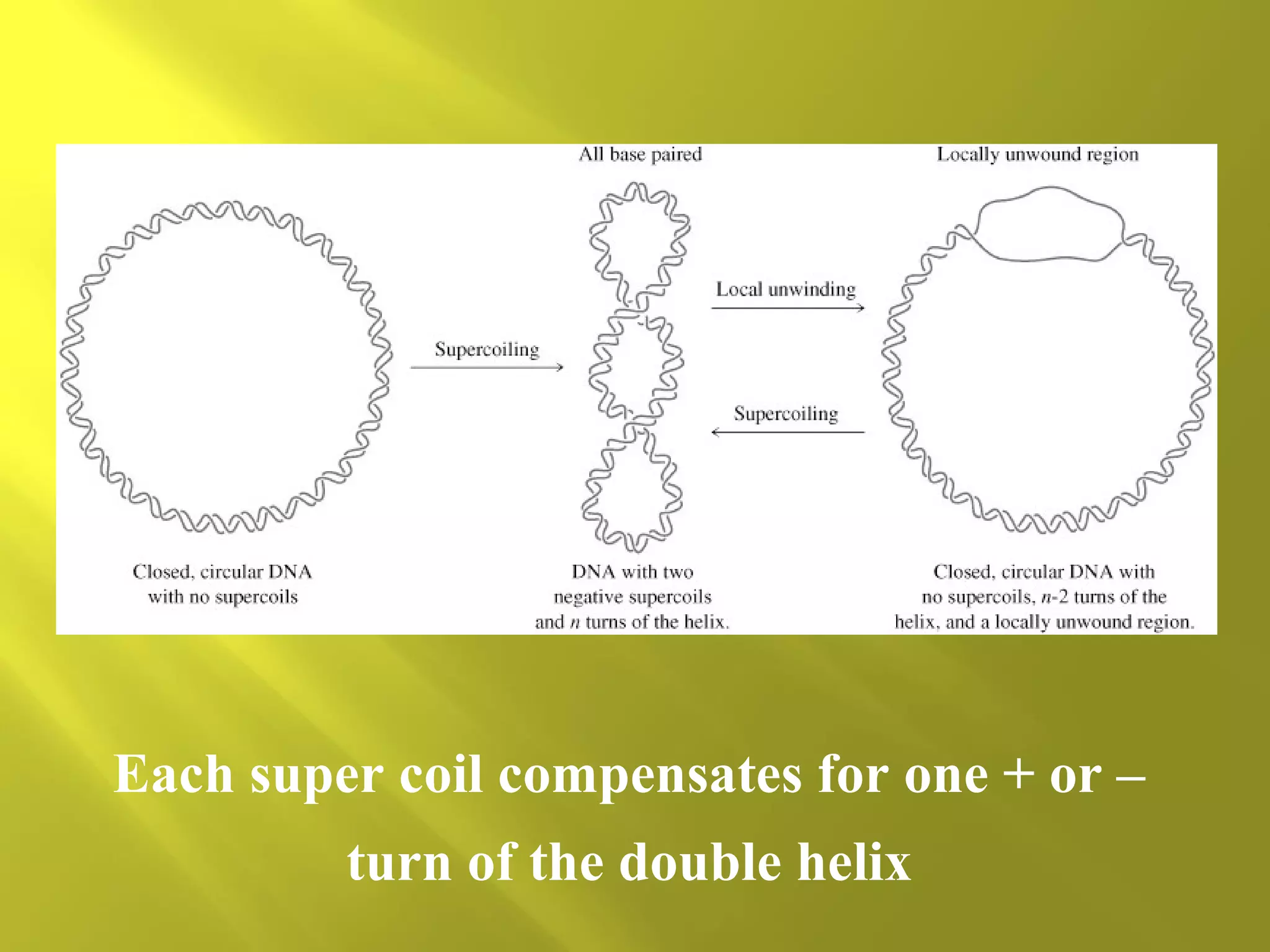
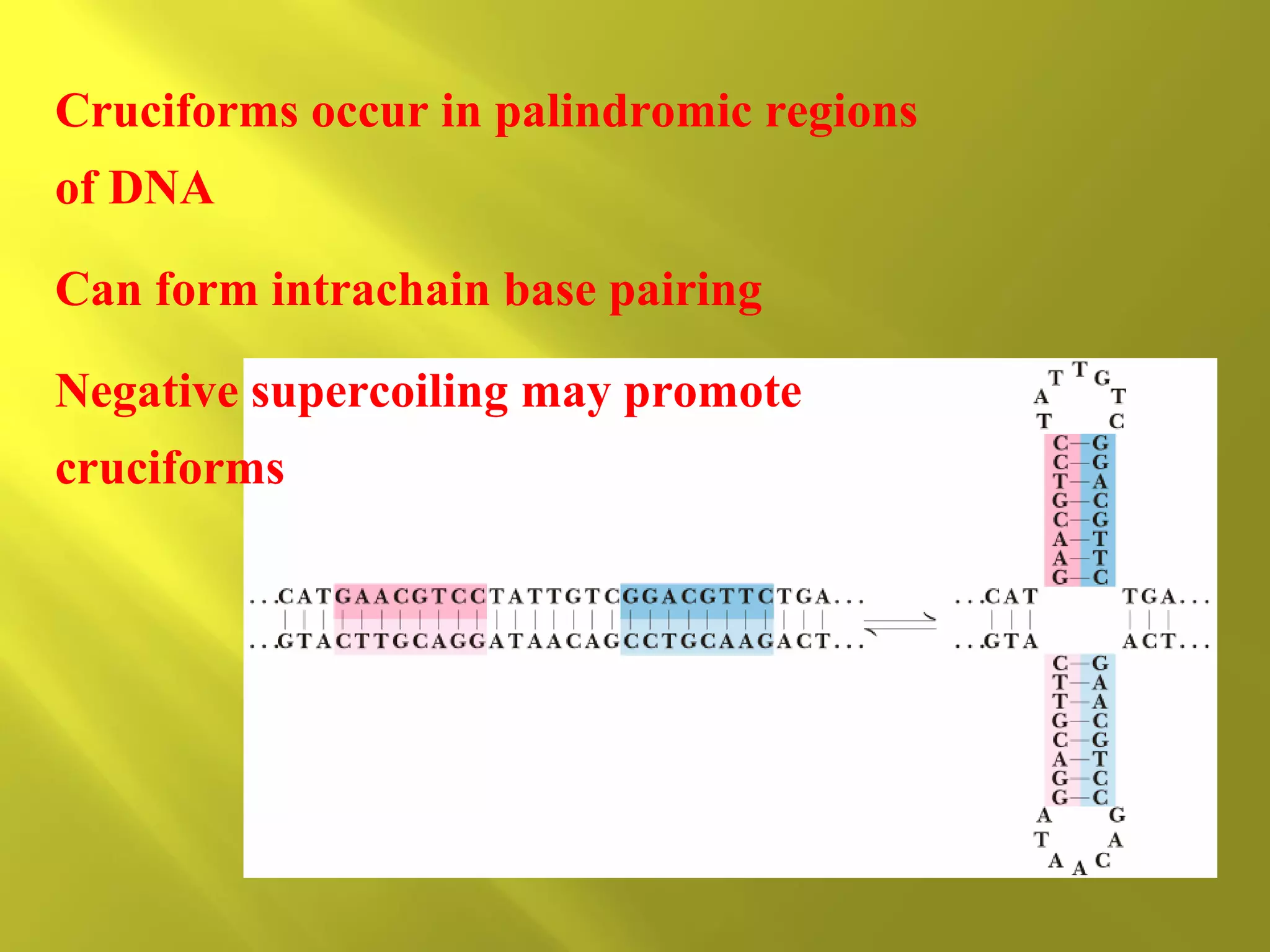
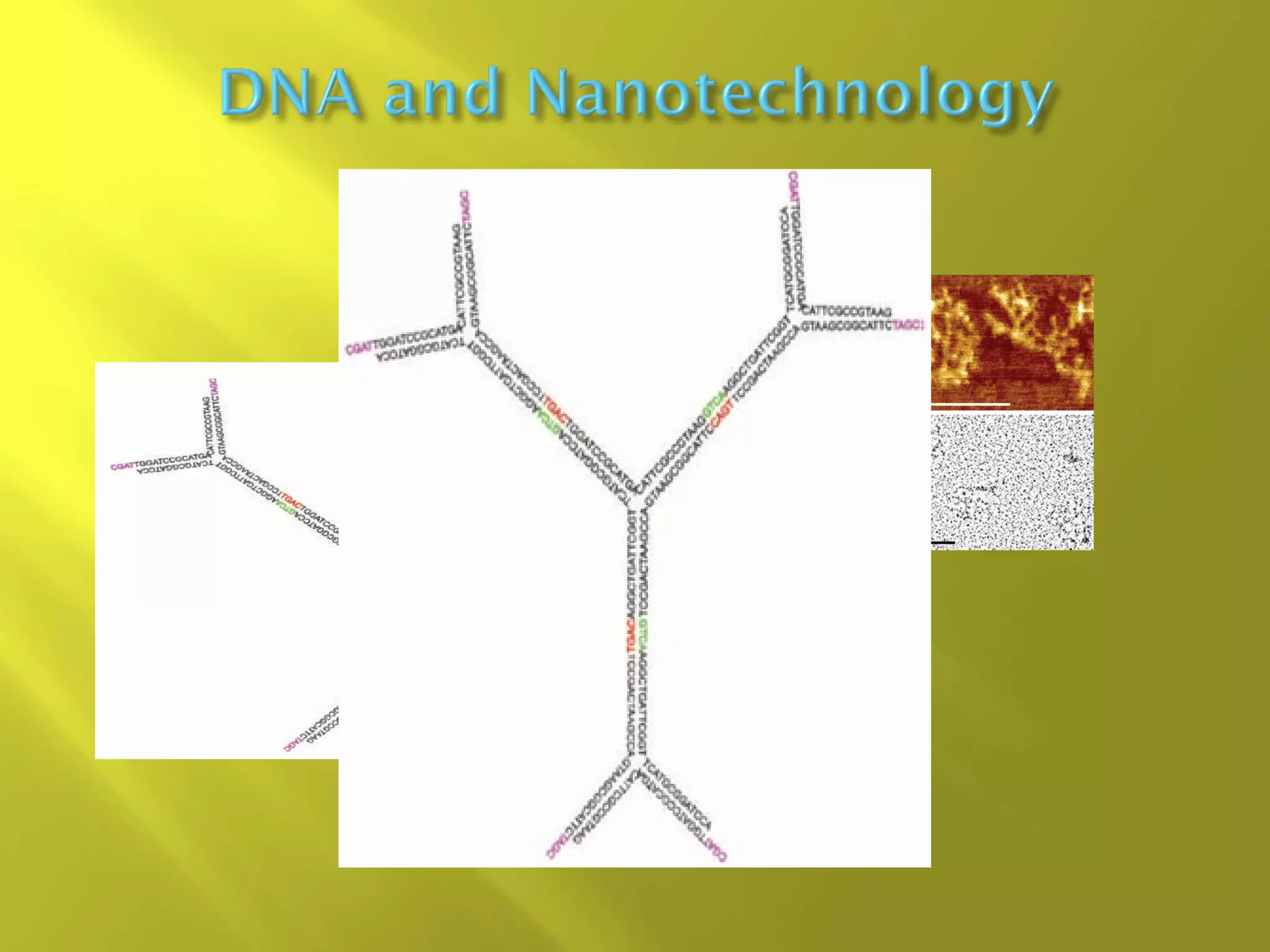
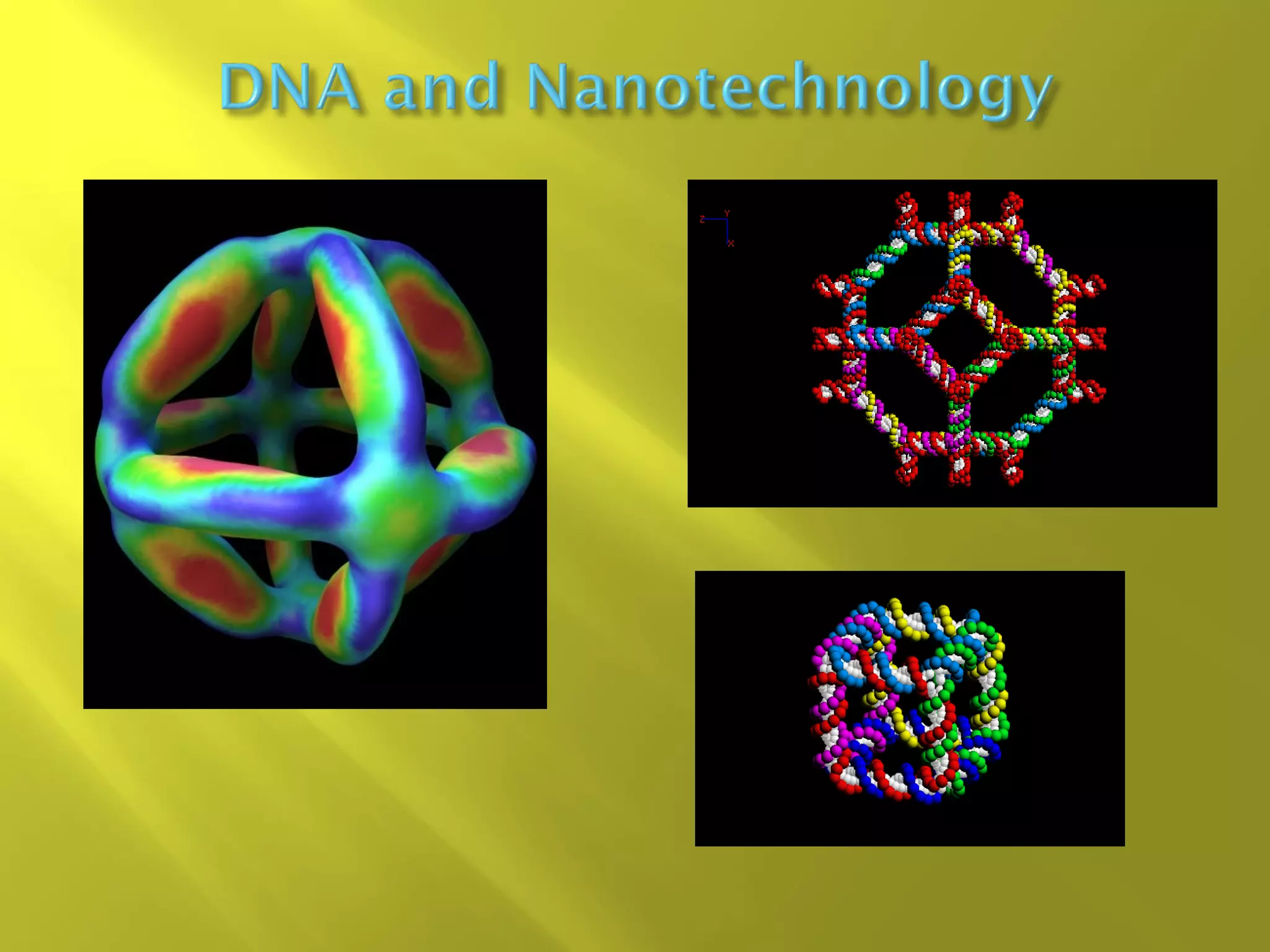
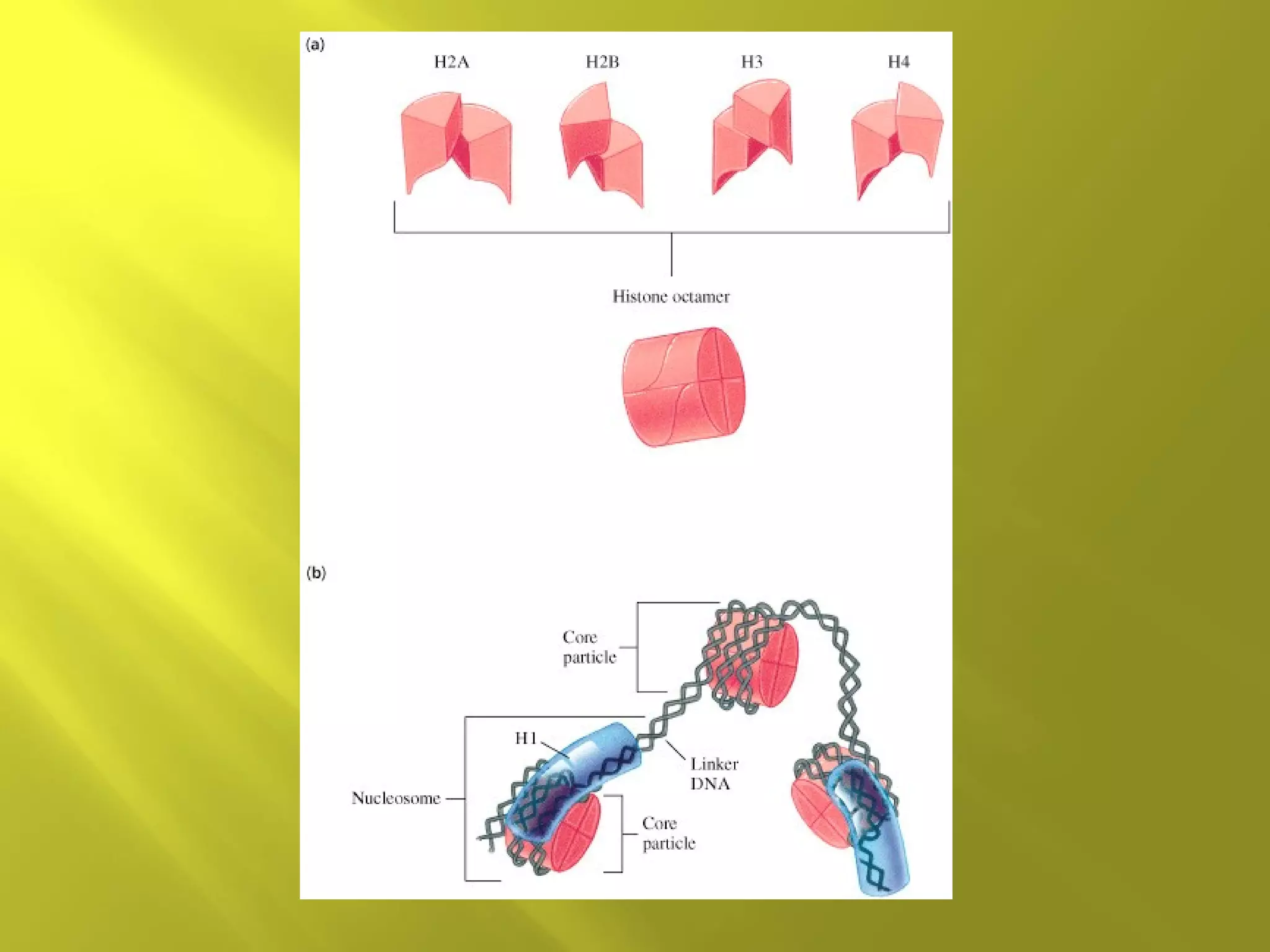
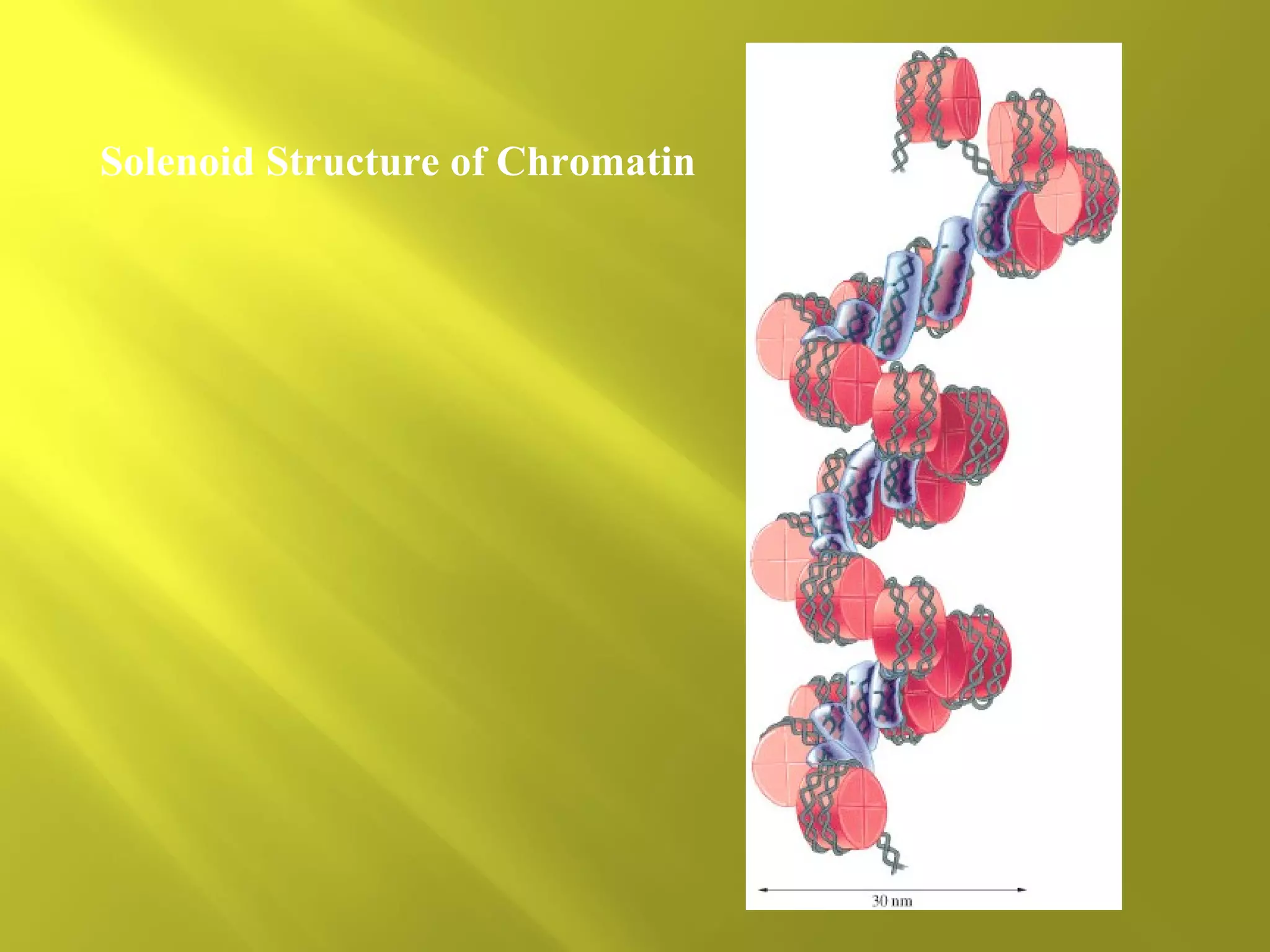
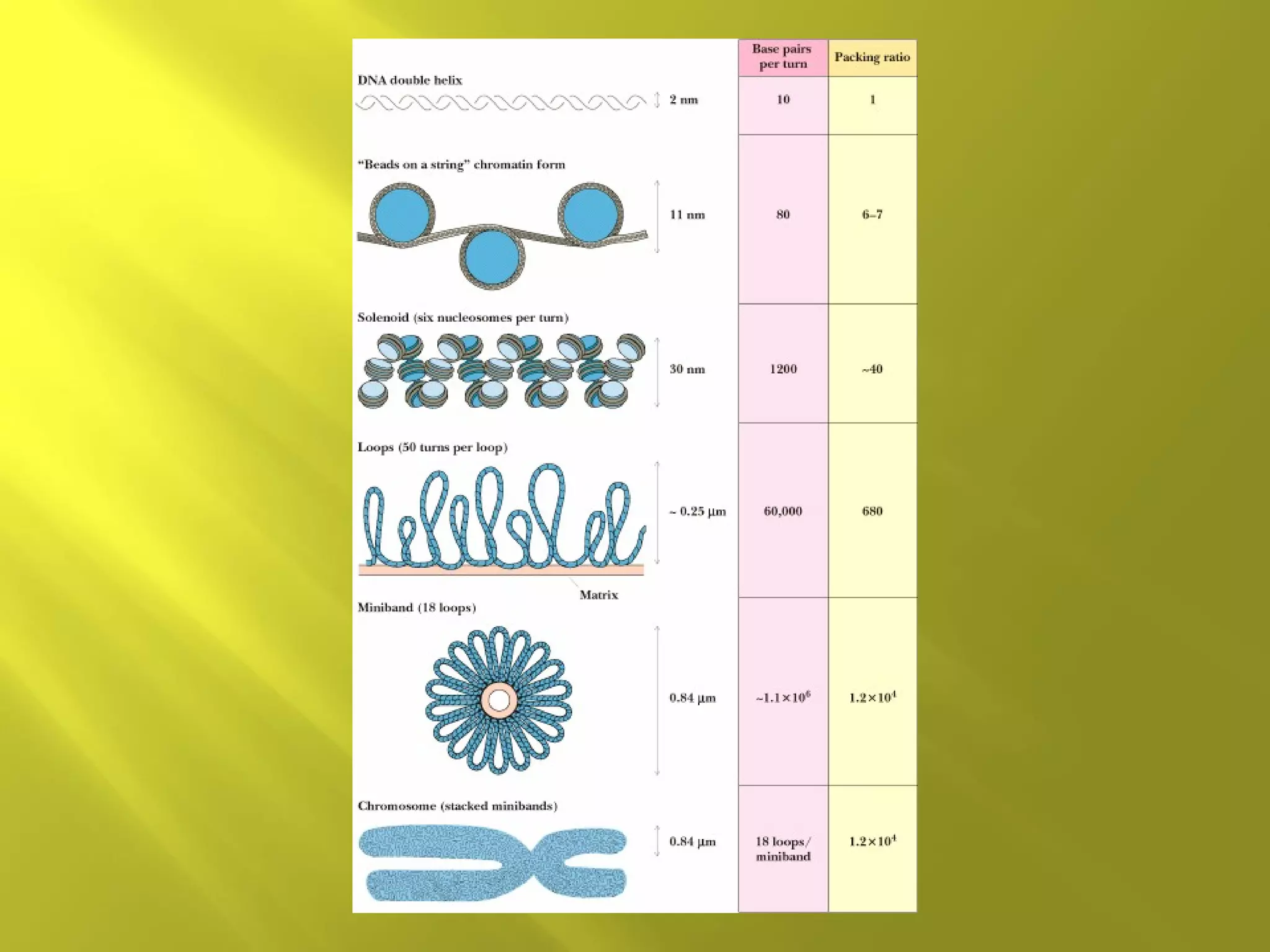
1. DNA structure involves four levels - from nucleotides to chromatin packaging. DNA sequencing methods like Sanger sequencing involve chain termination using dideoxynucleotides.
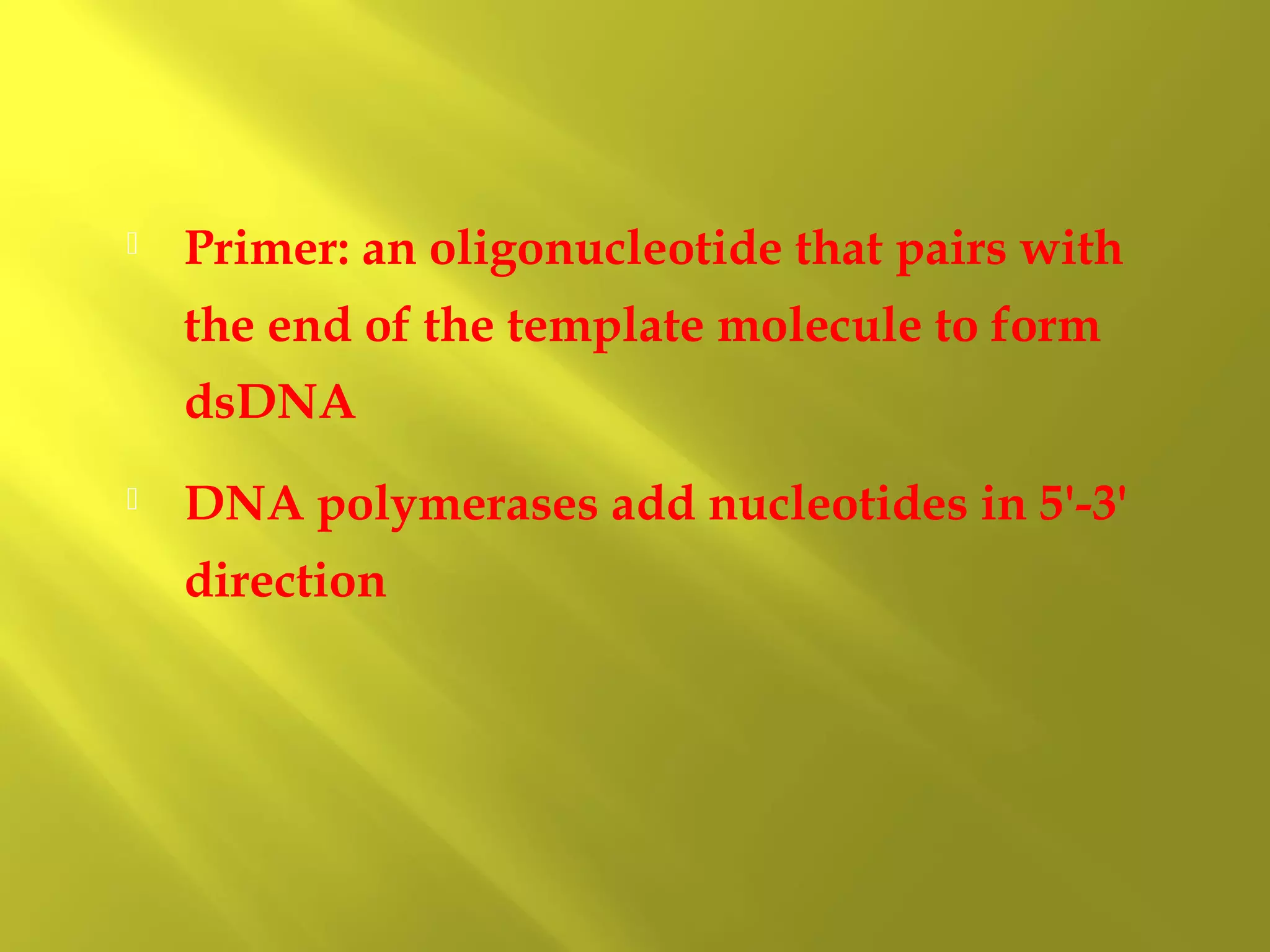
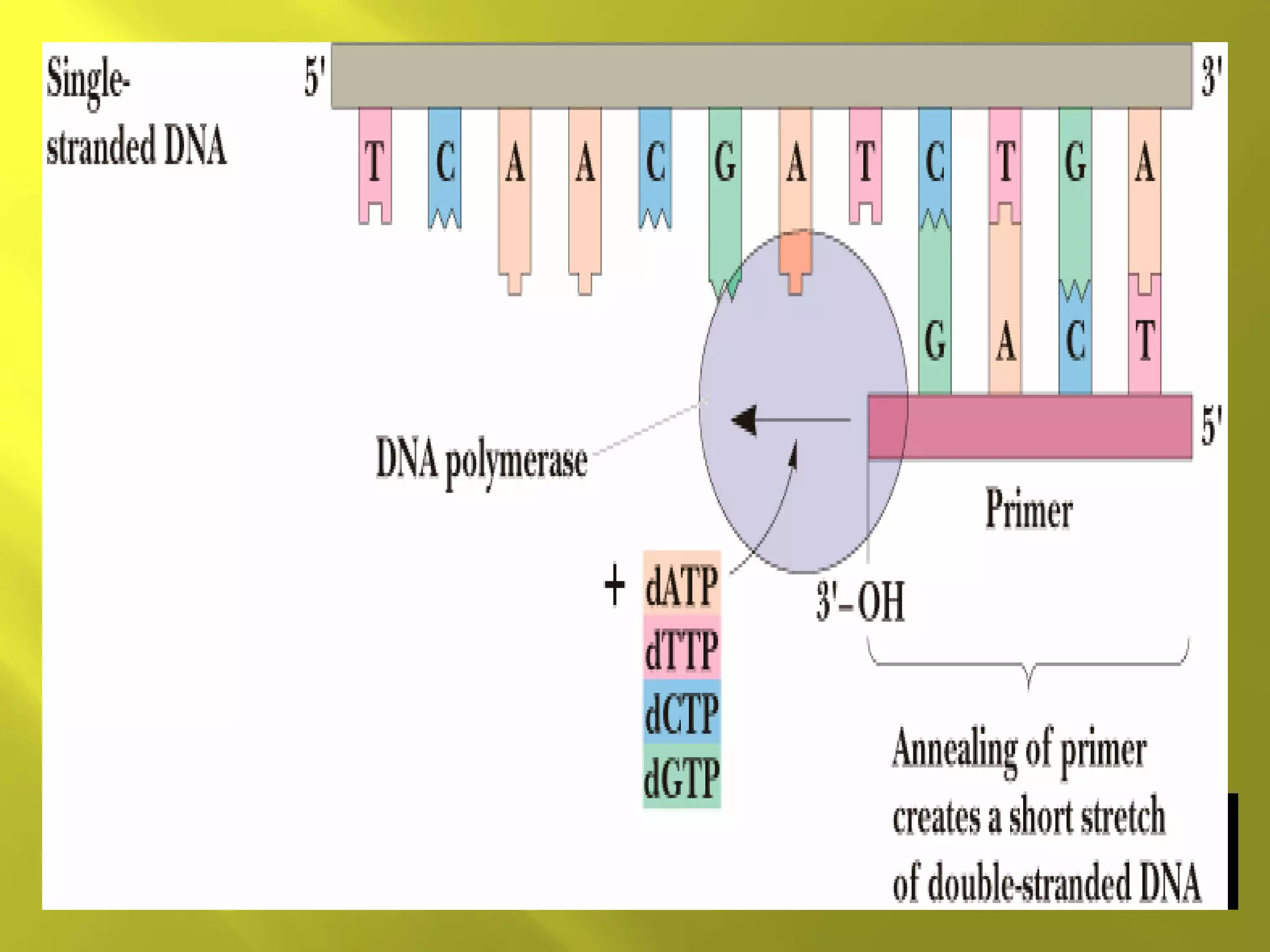
2. DNA replication requires primers, DNA polymerase and occurs in 5' to 3' direction on each strand.
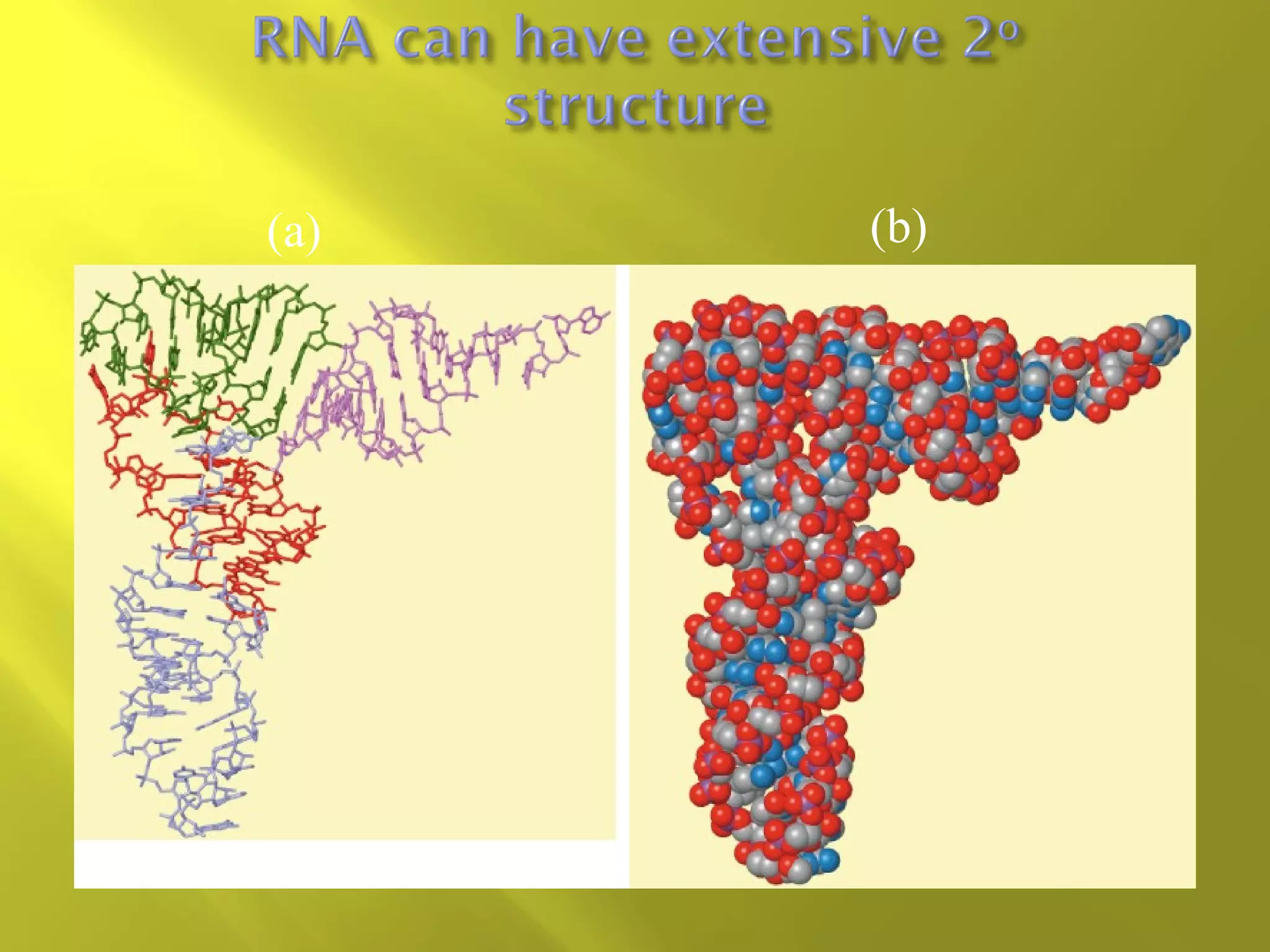
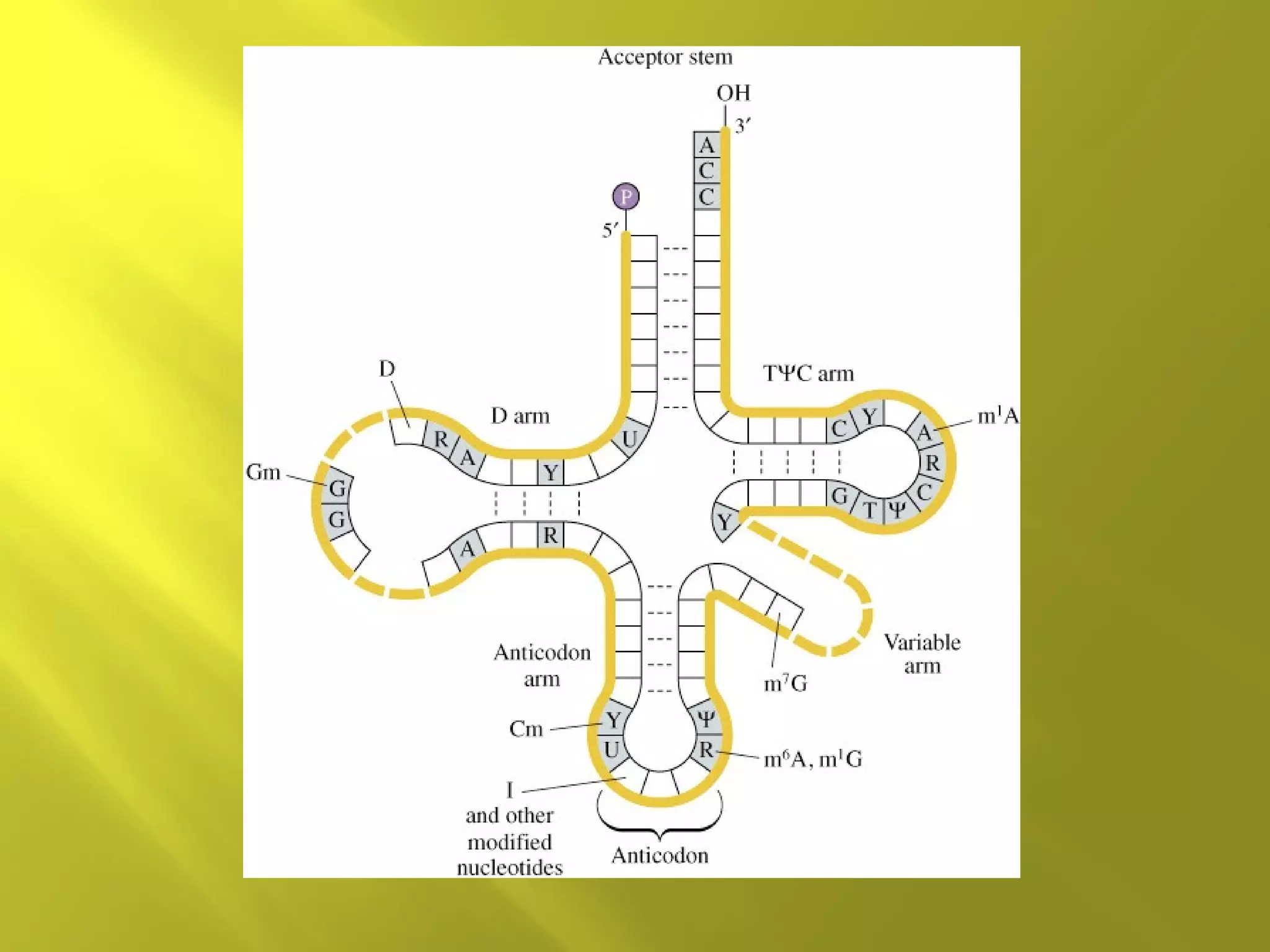
3. RNA differs from DNA in being single-stranded and more chemically unstable but has important functions as rRNA, tRNA, mRNA and ribozymes.