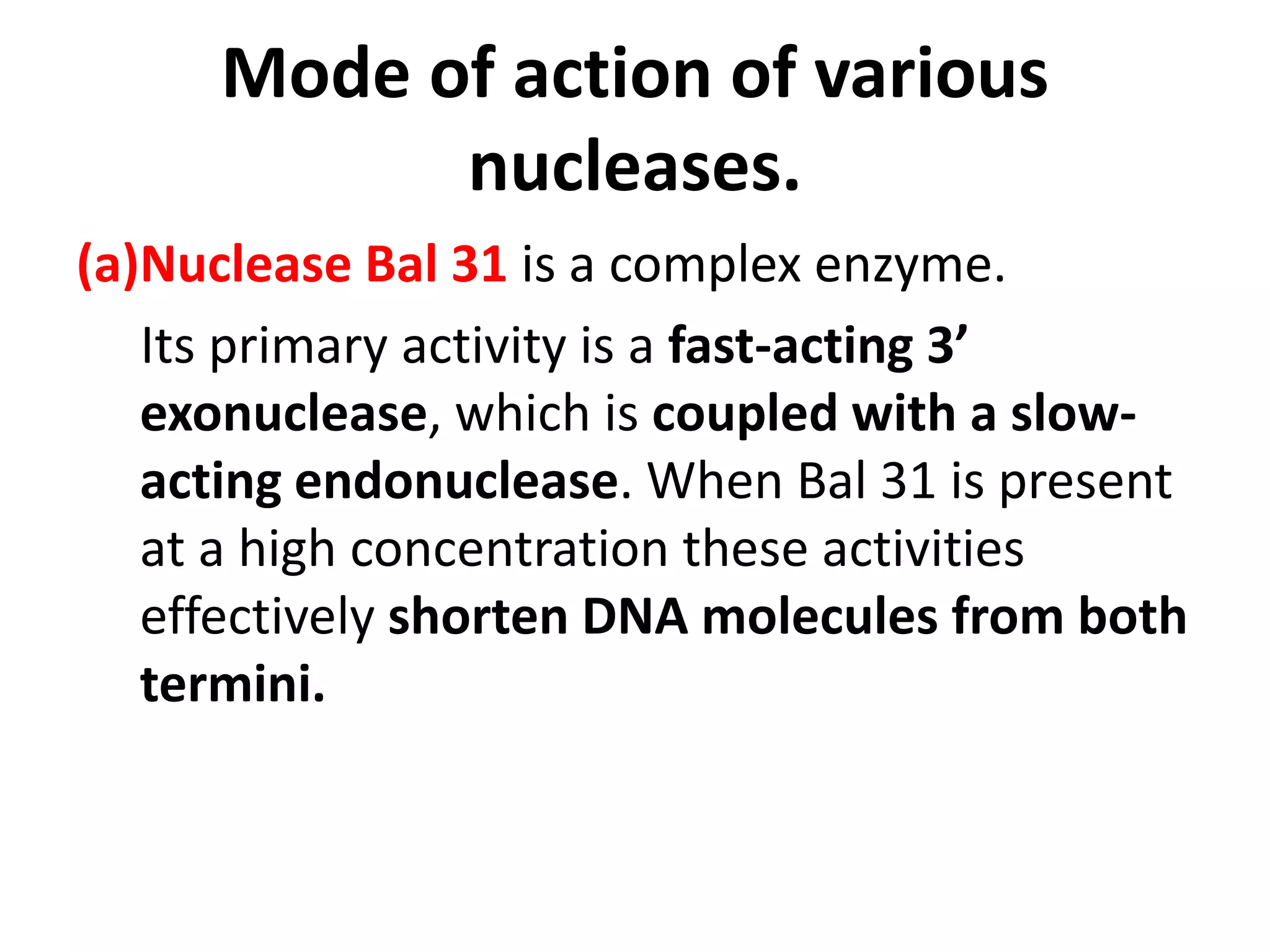
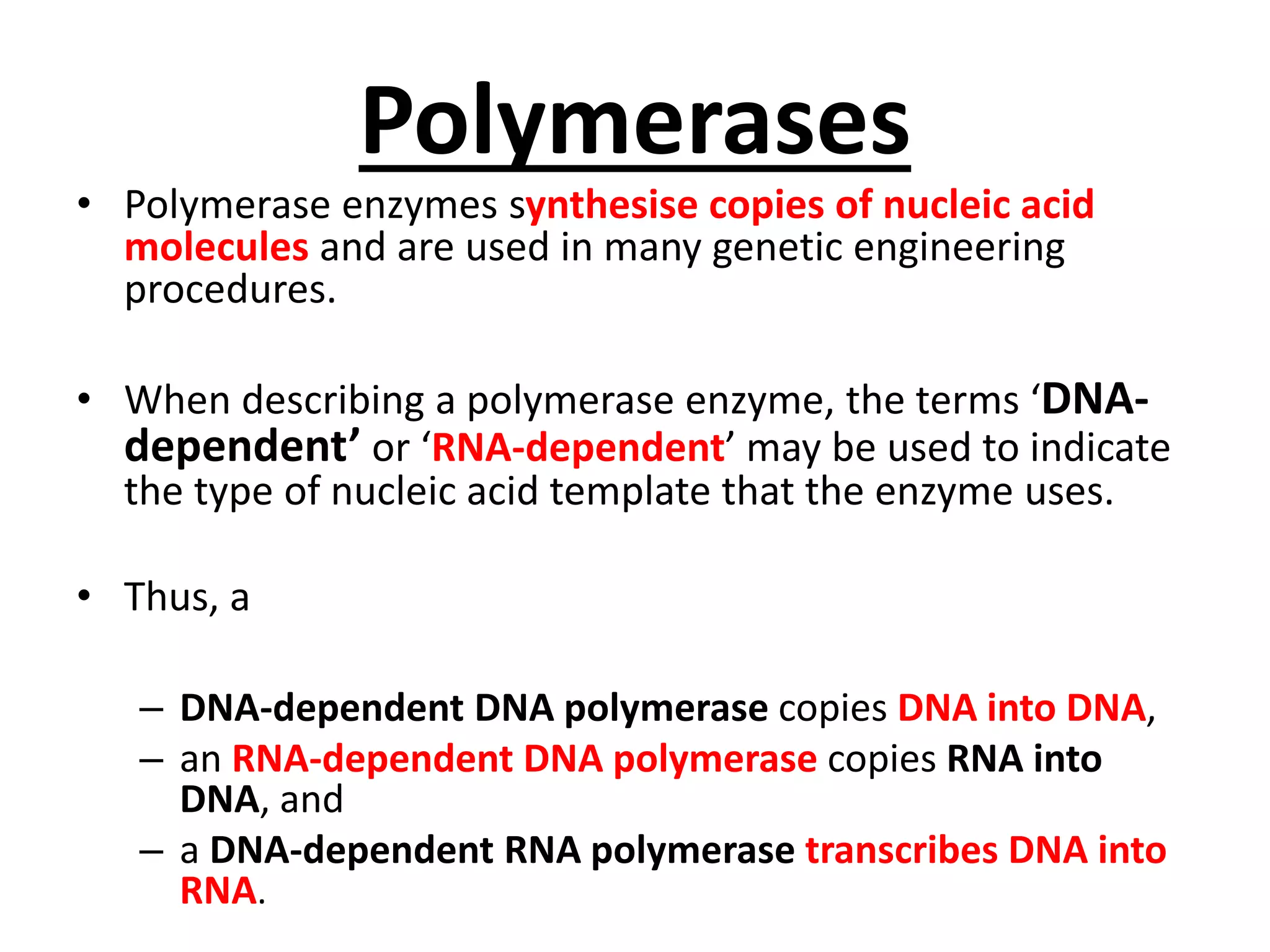
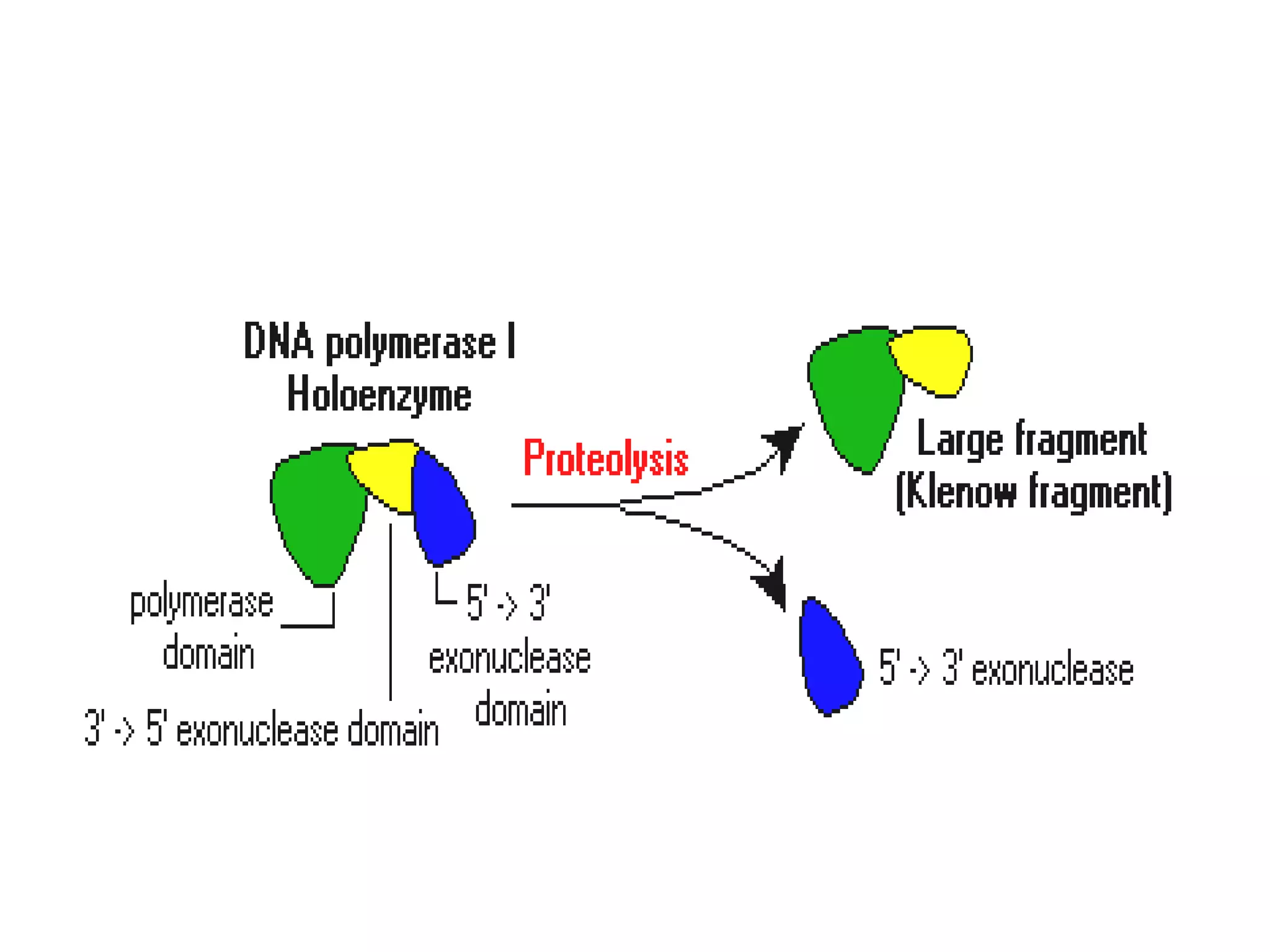
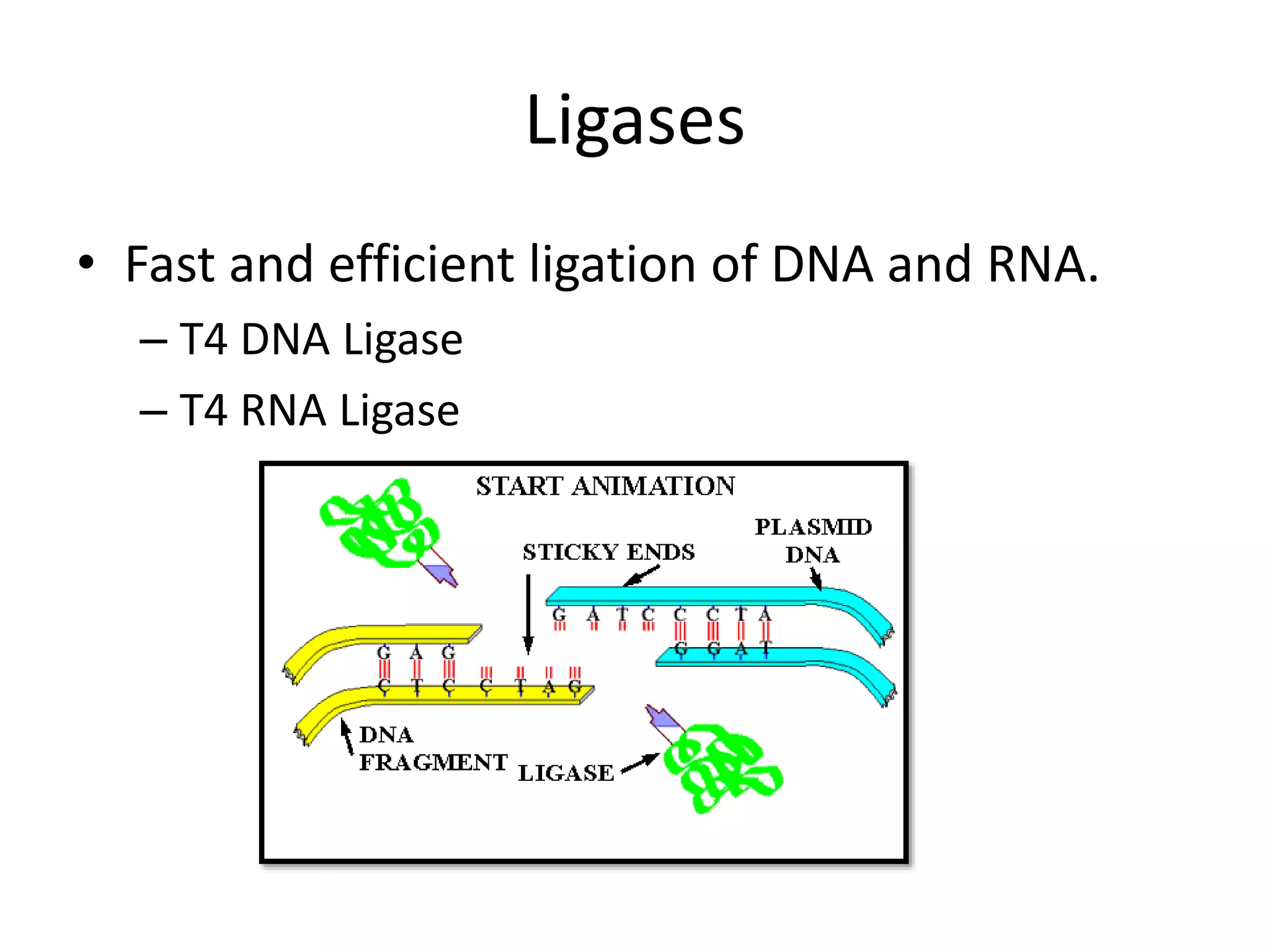
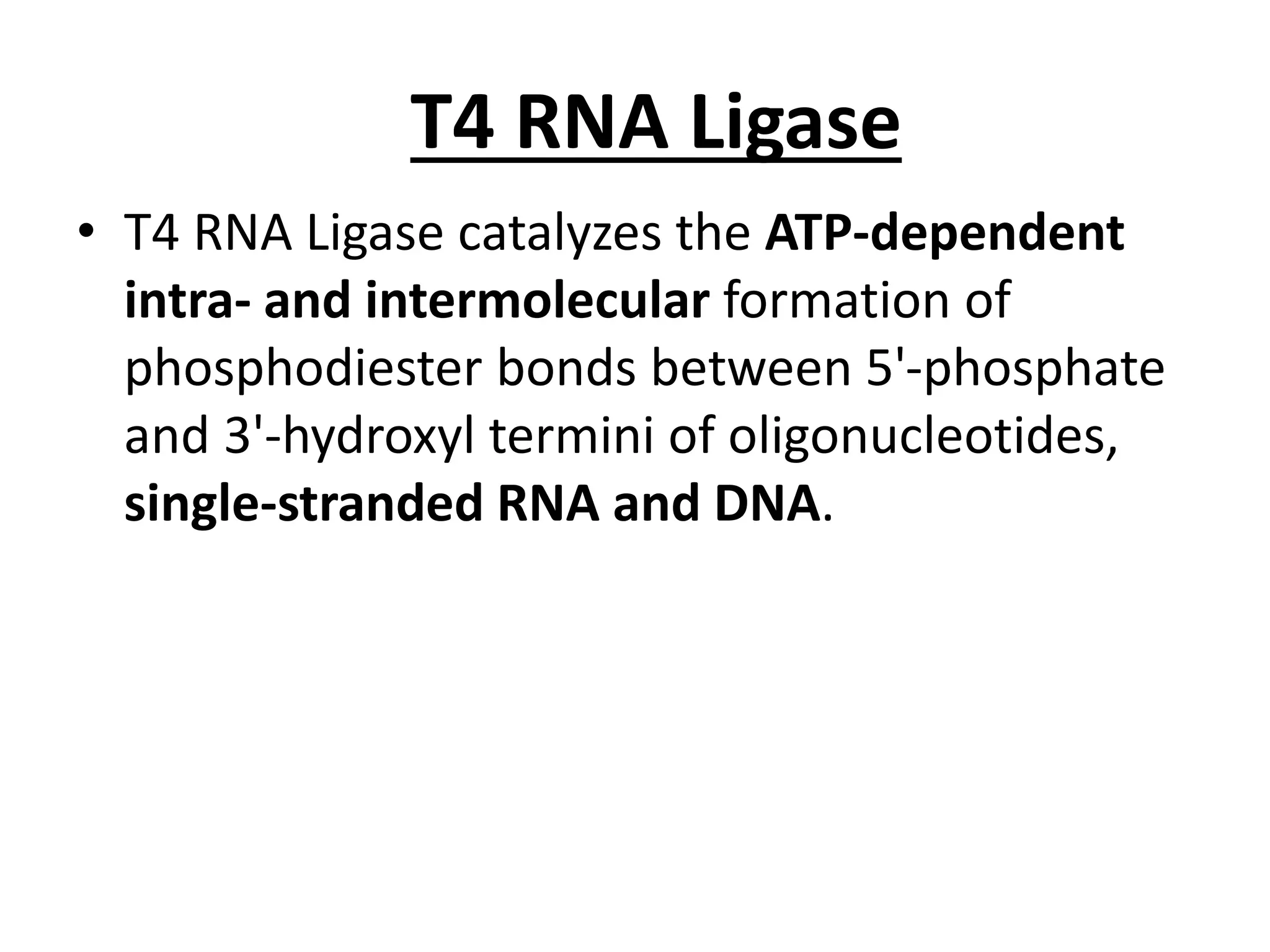
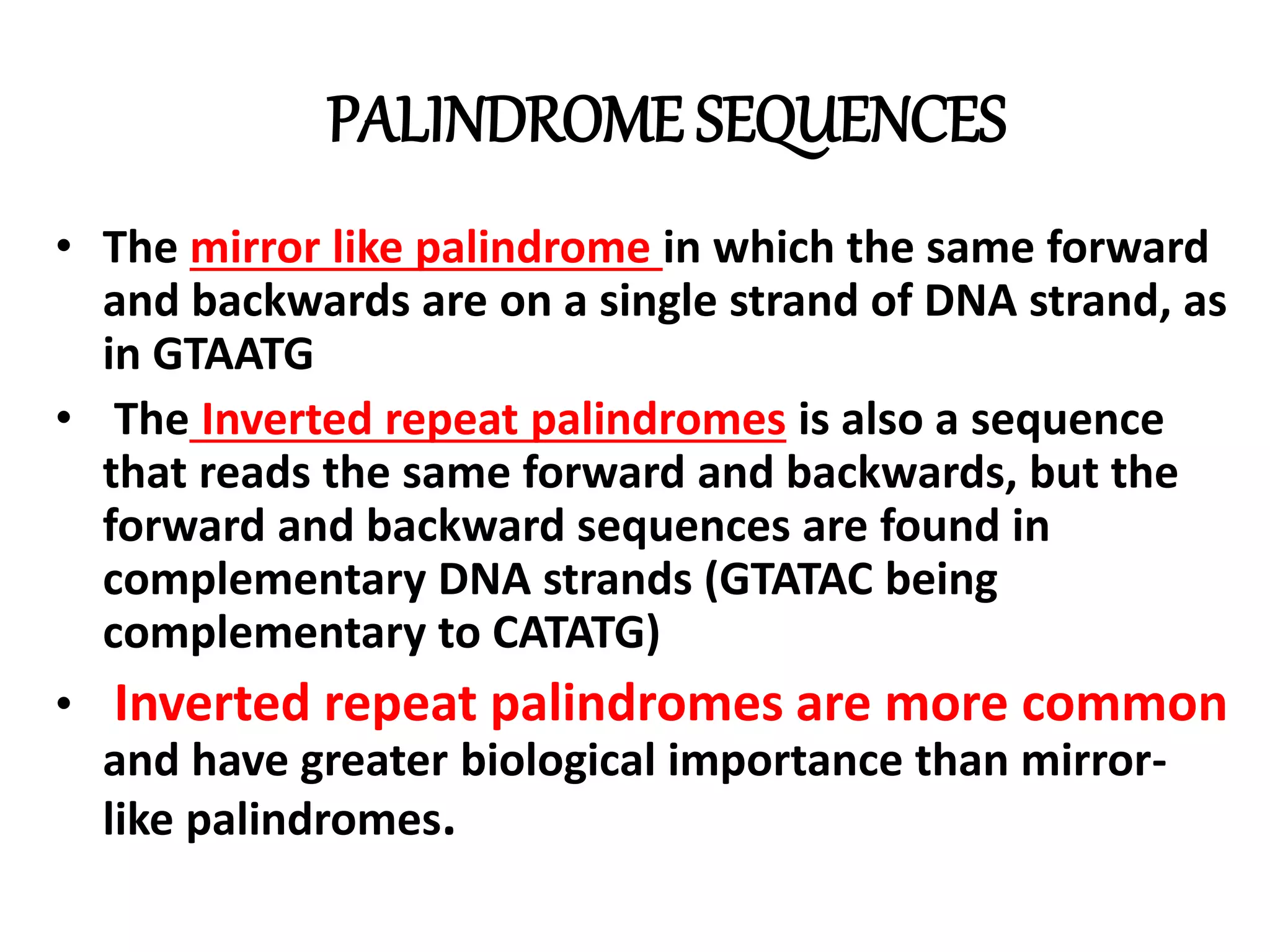
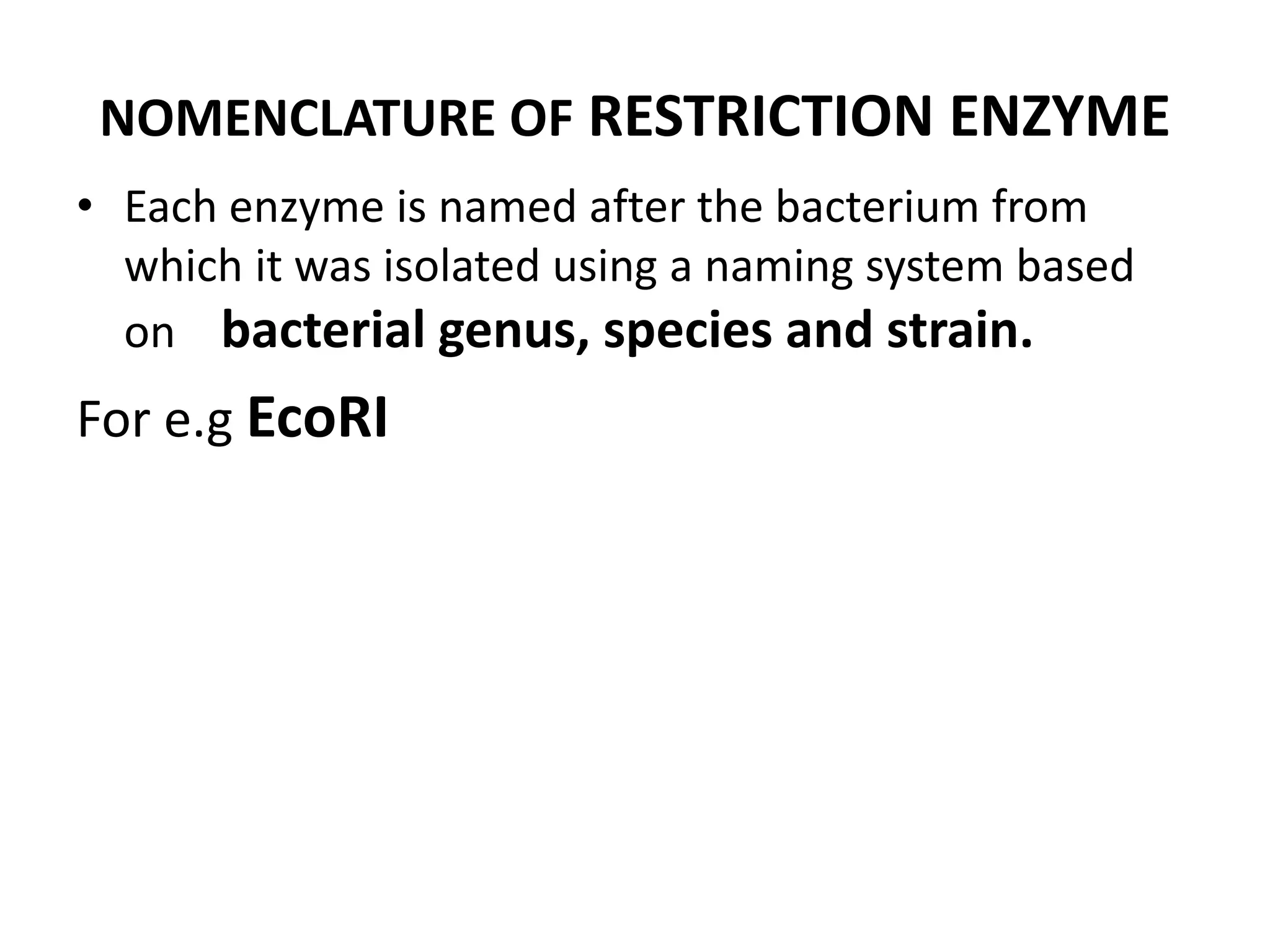
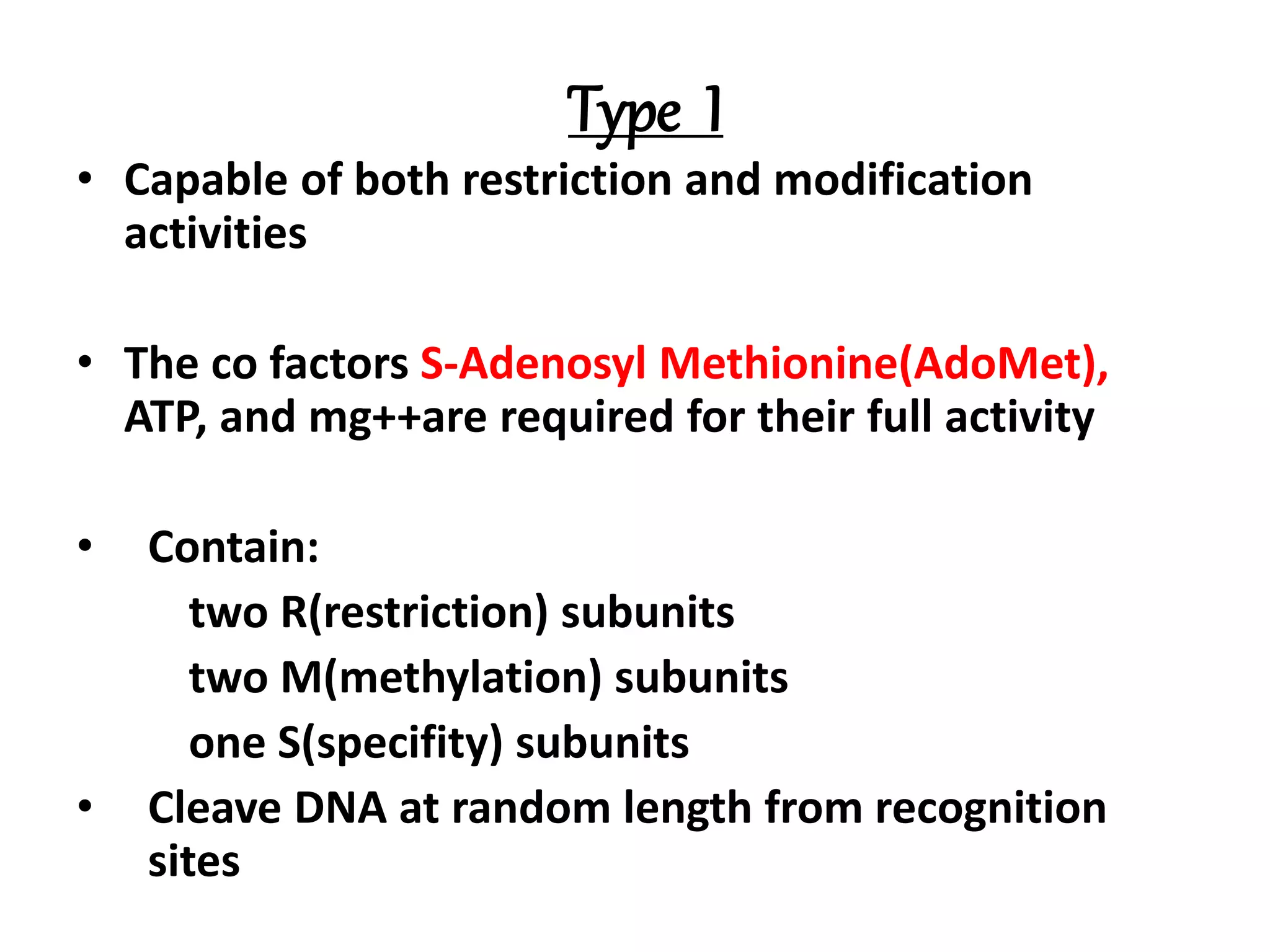
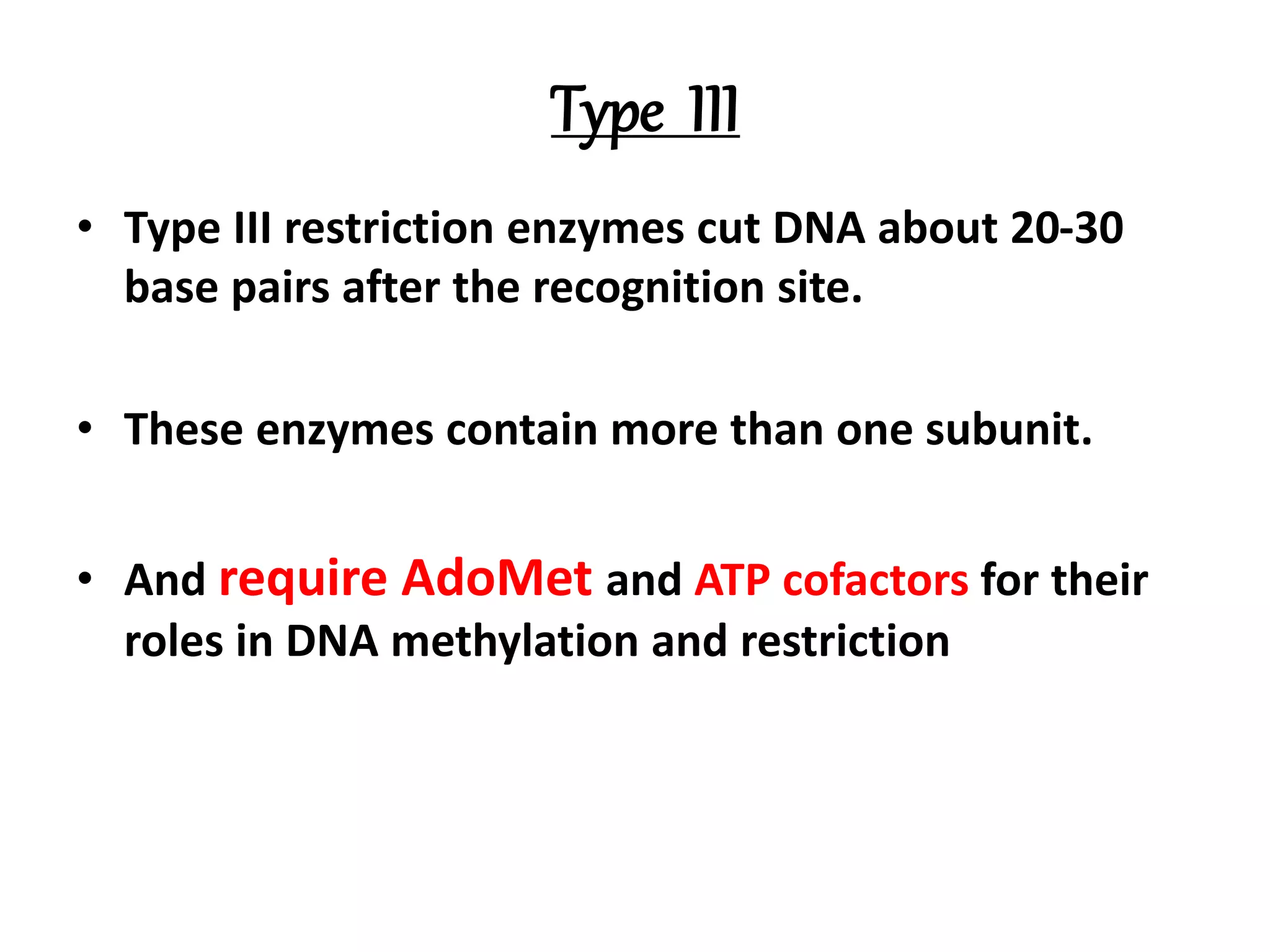
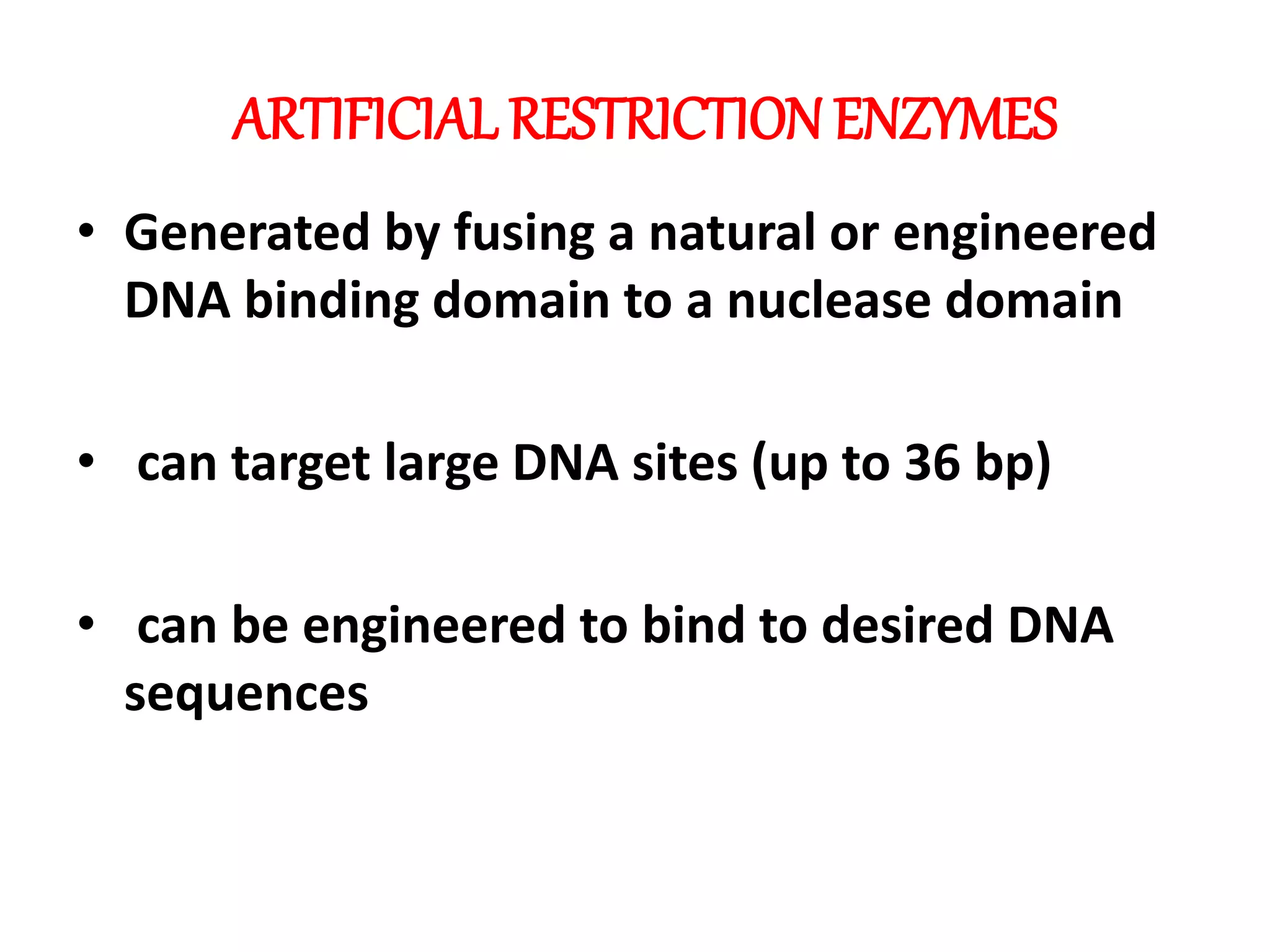
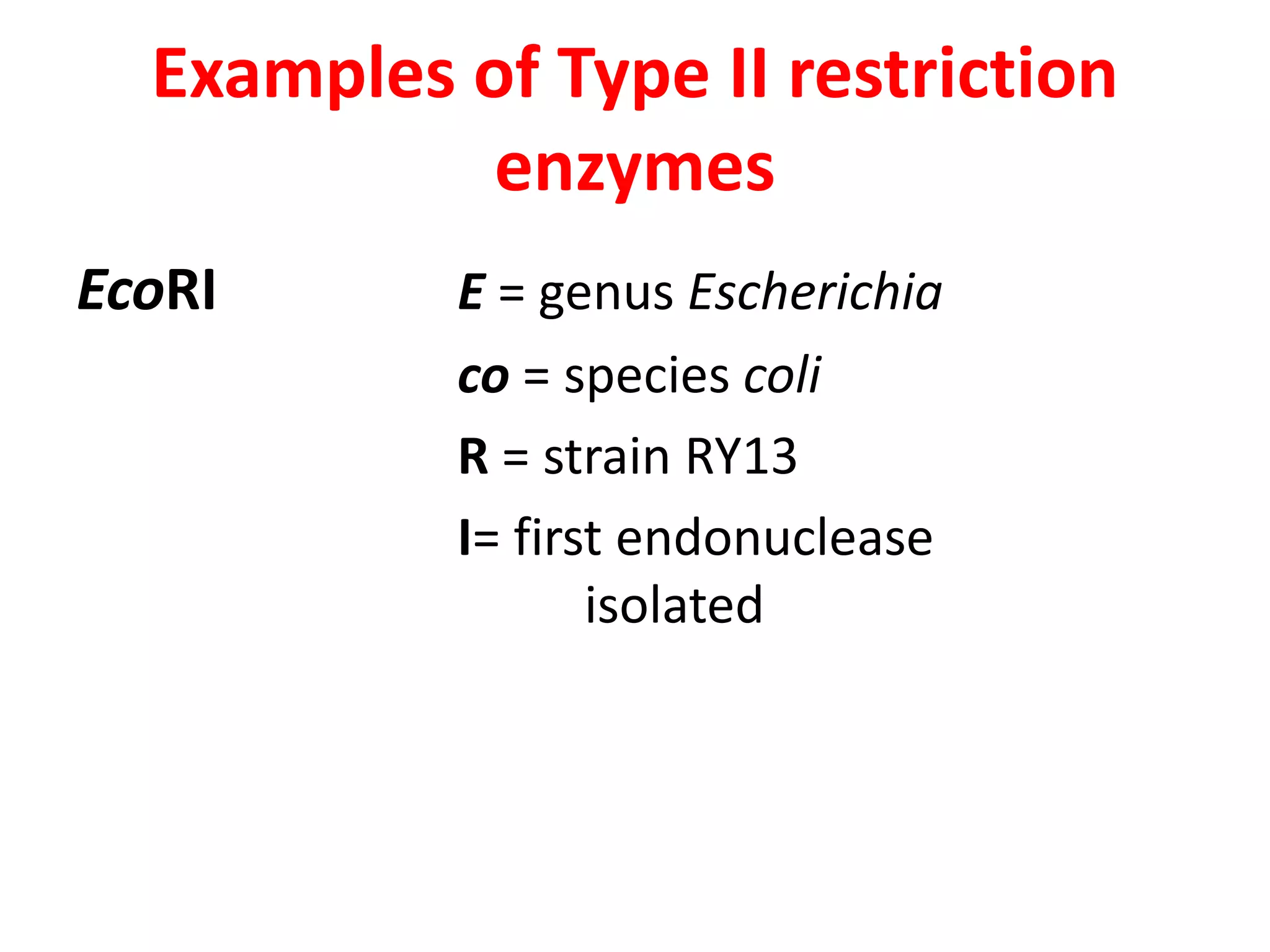
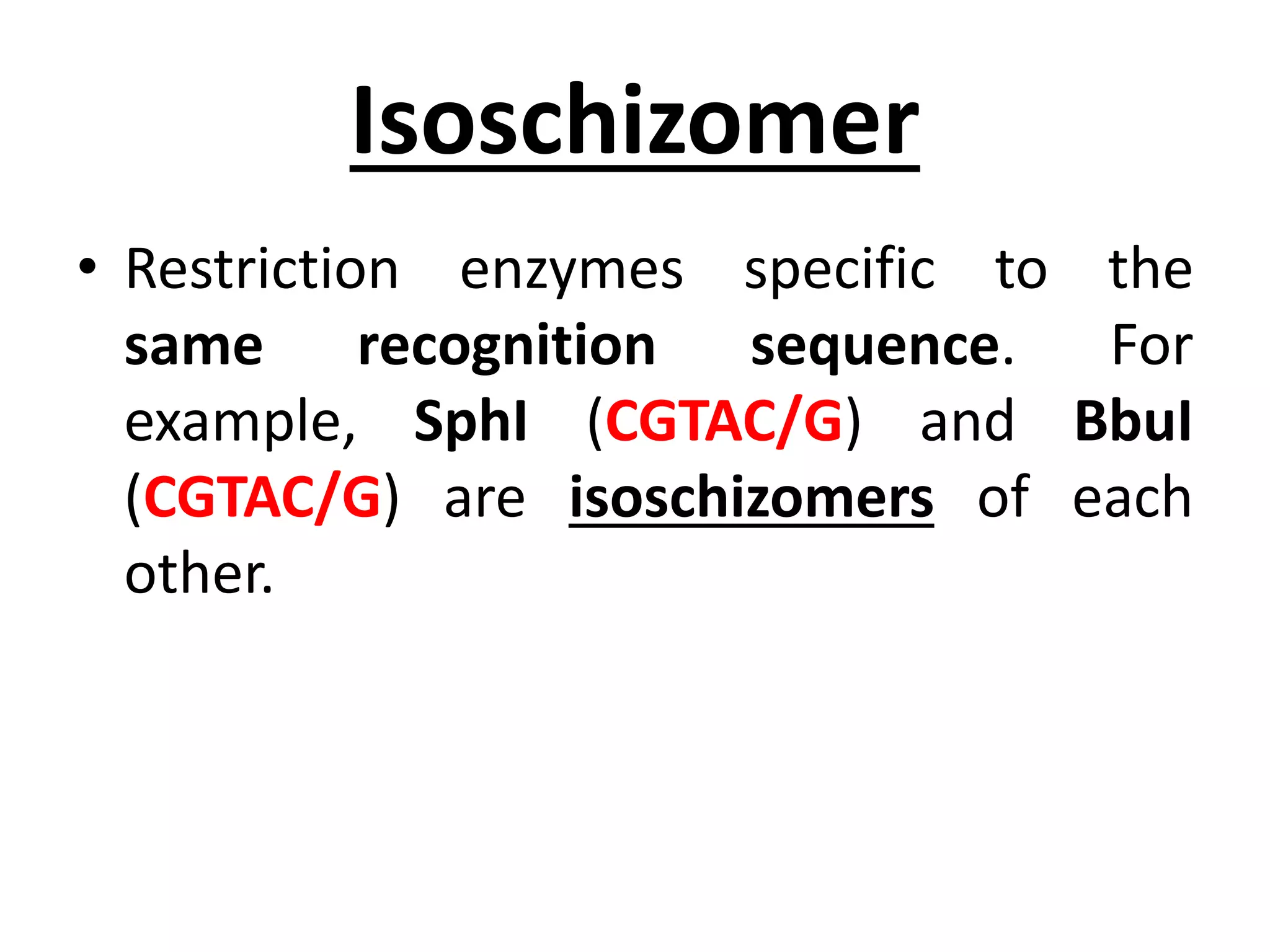
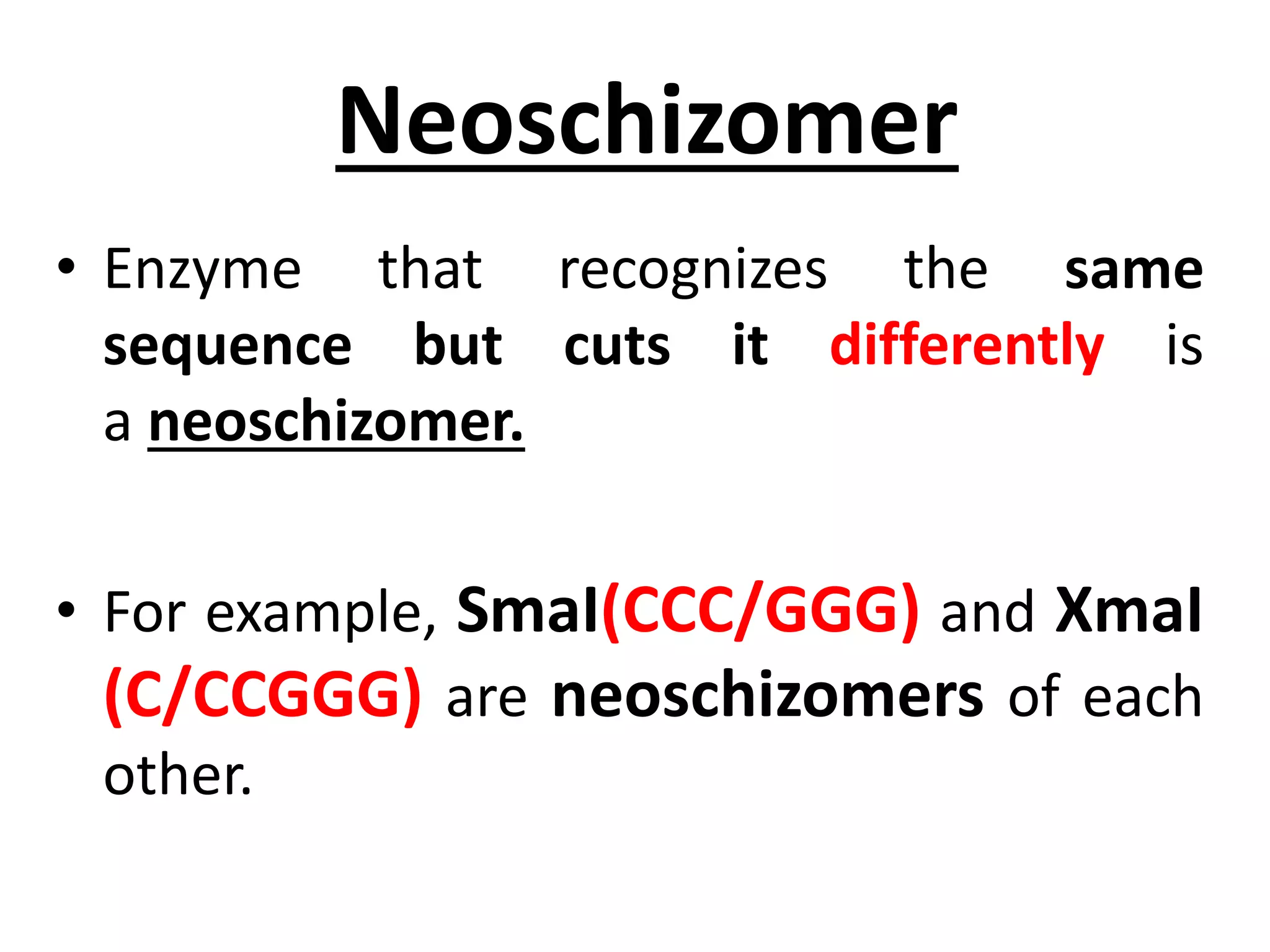
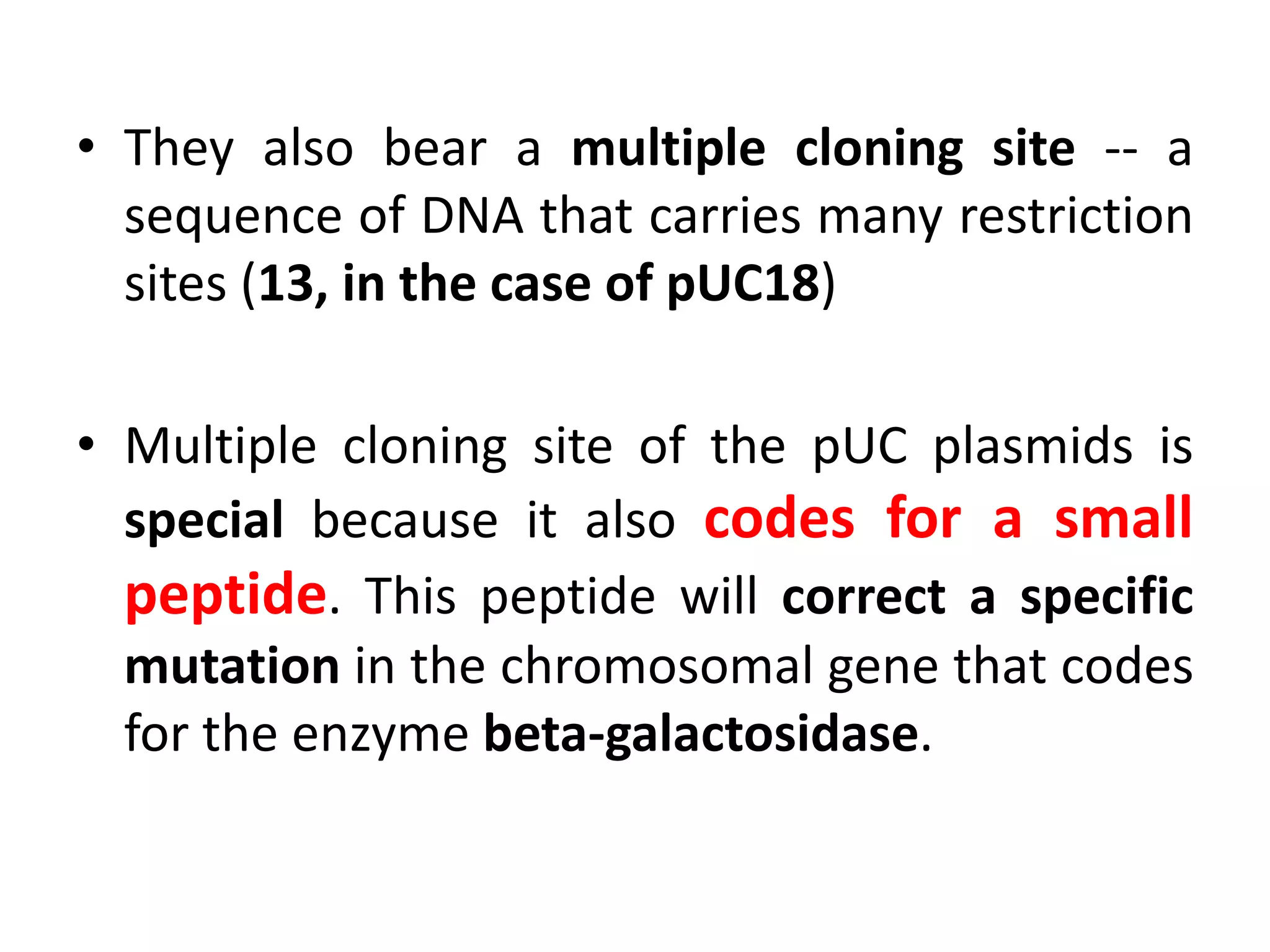
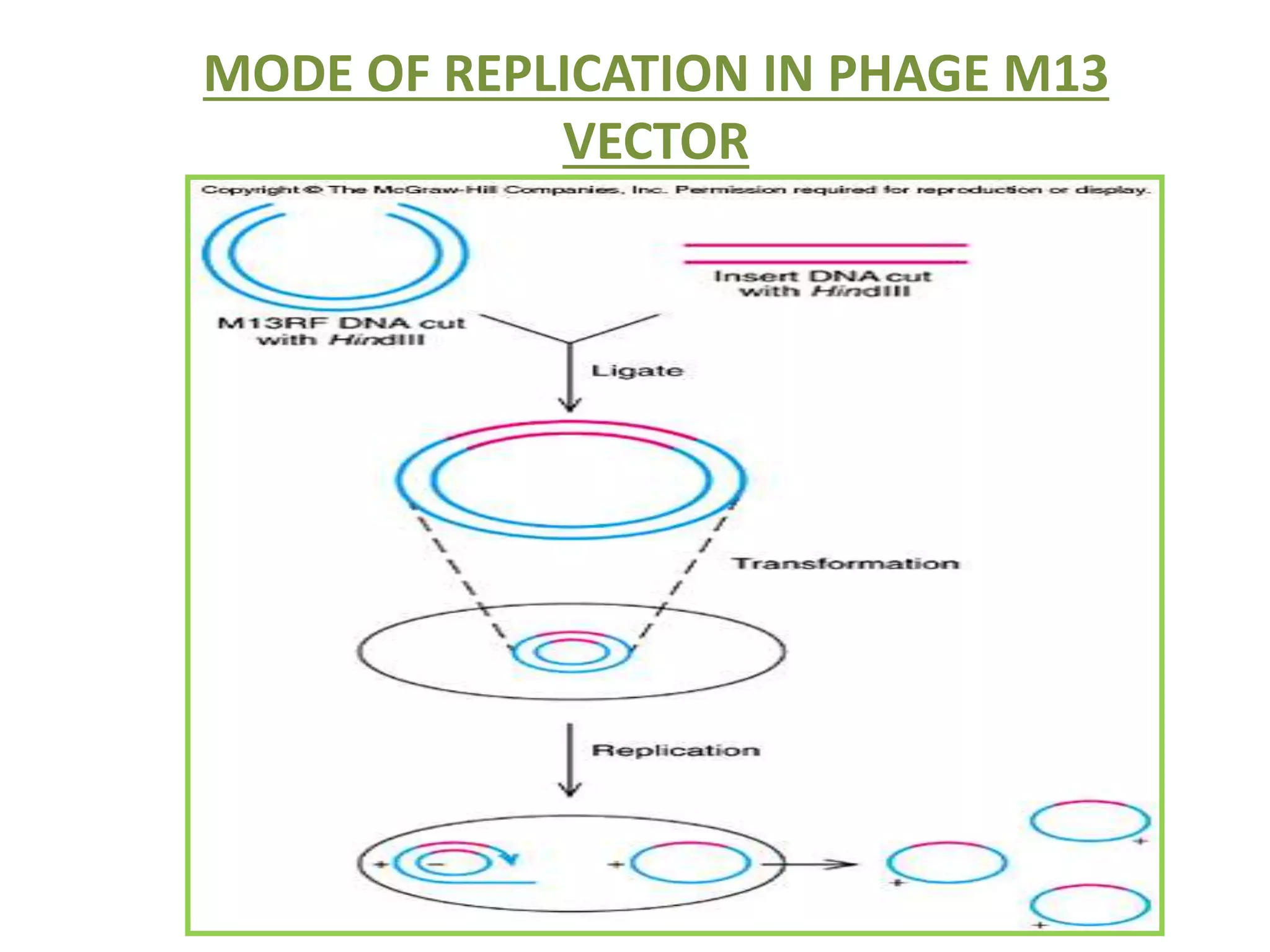
This document discusses enzymes used in genetic engineering, specifically focusing on restriction enzymes and DNA modifying enzymes. It provides details on various types of modifying enzymes including nucleases, polymerases, phosphatases, kinases, ligases and others. Restriction enzymes are described as molecular scissors that cut DNA at specific recognition sequences. DNA ligase is presented as the molecular glue that joins cut DNA fragments. The document outlines the classification, nomenclature, mechanisms and applications of various restriction enzymes and modifying enzymes used in genetic engineering techniques.