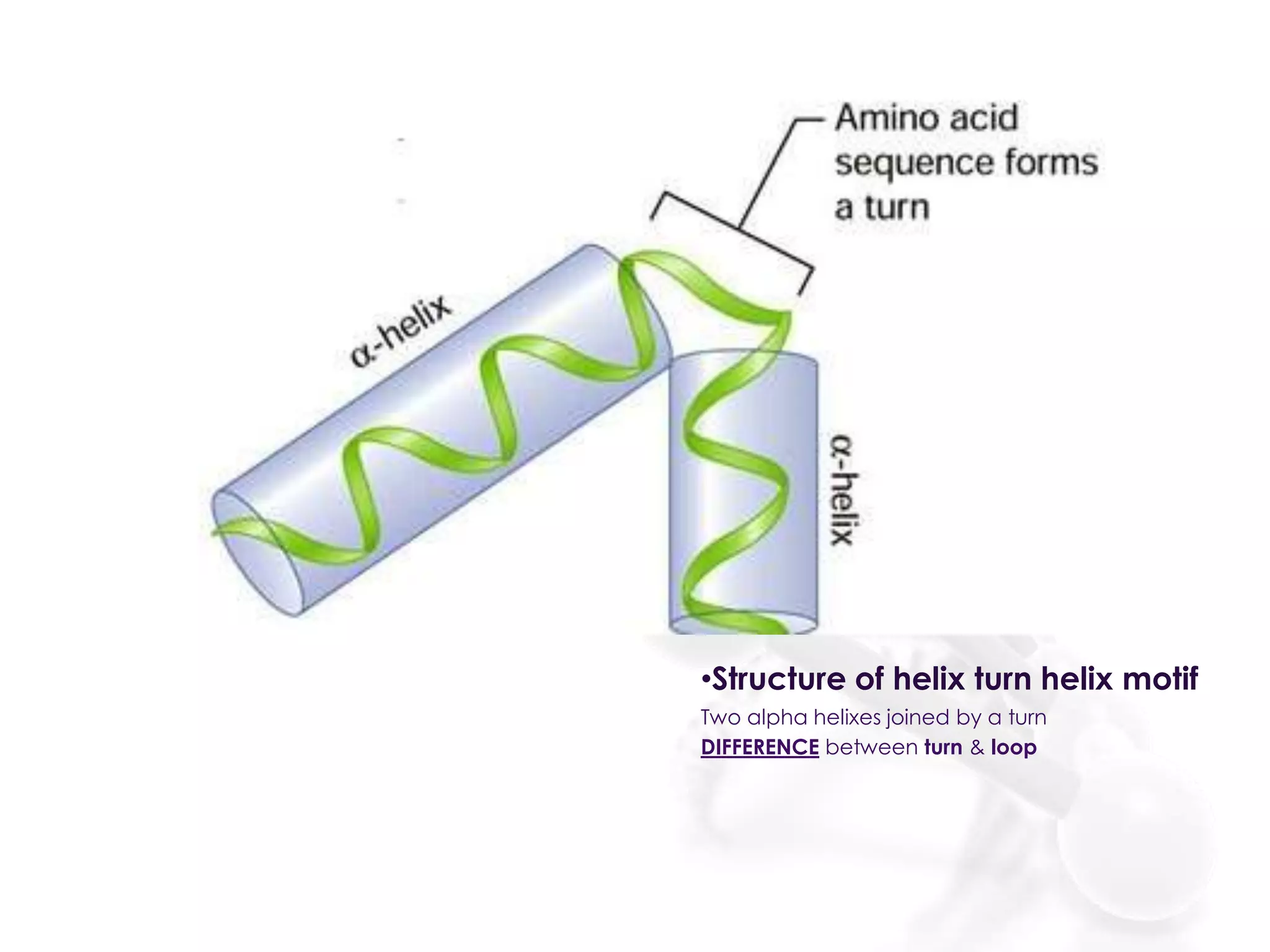
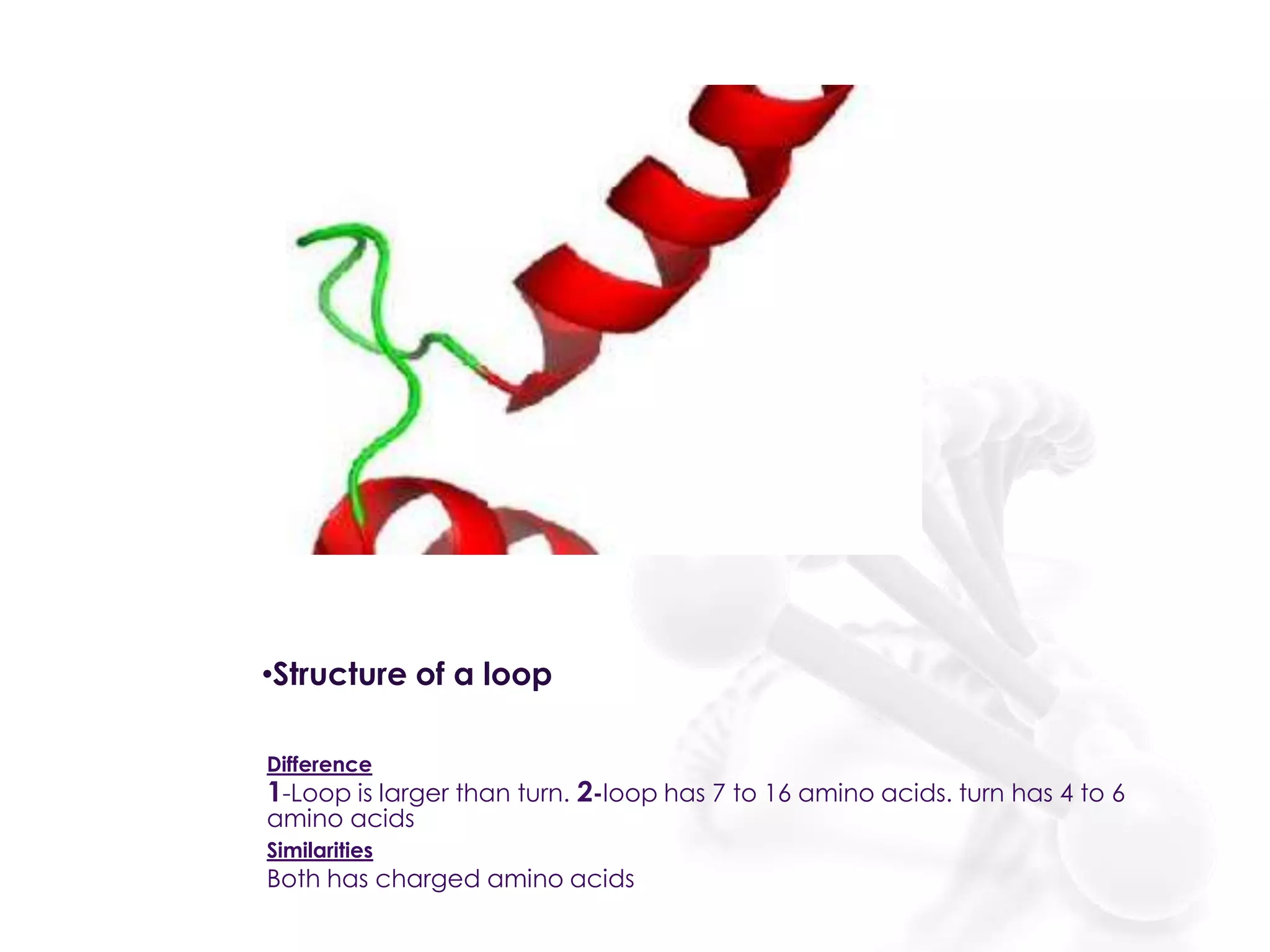
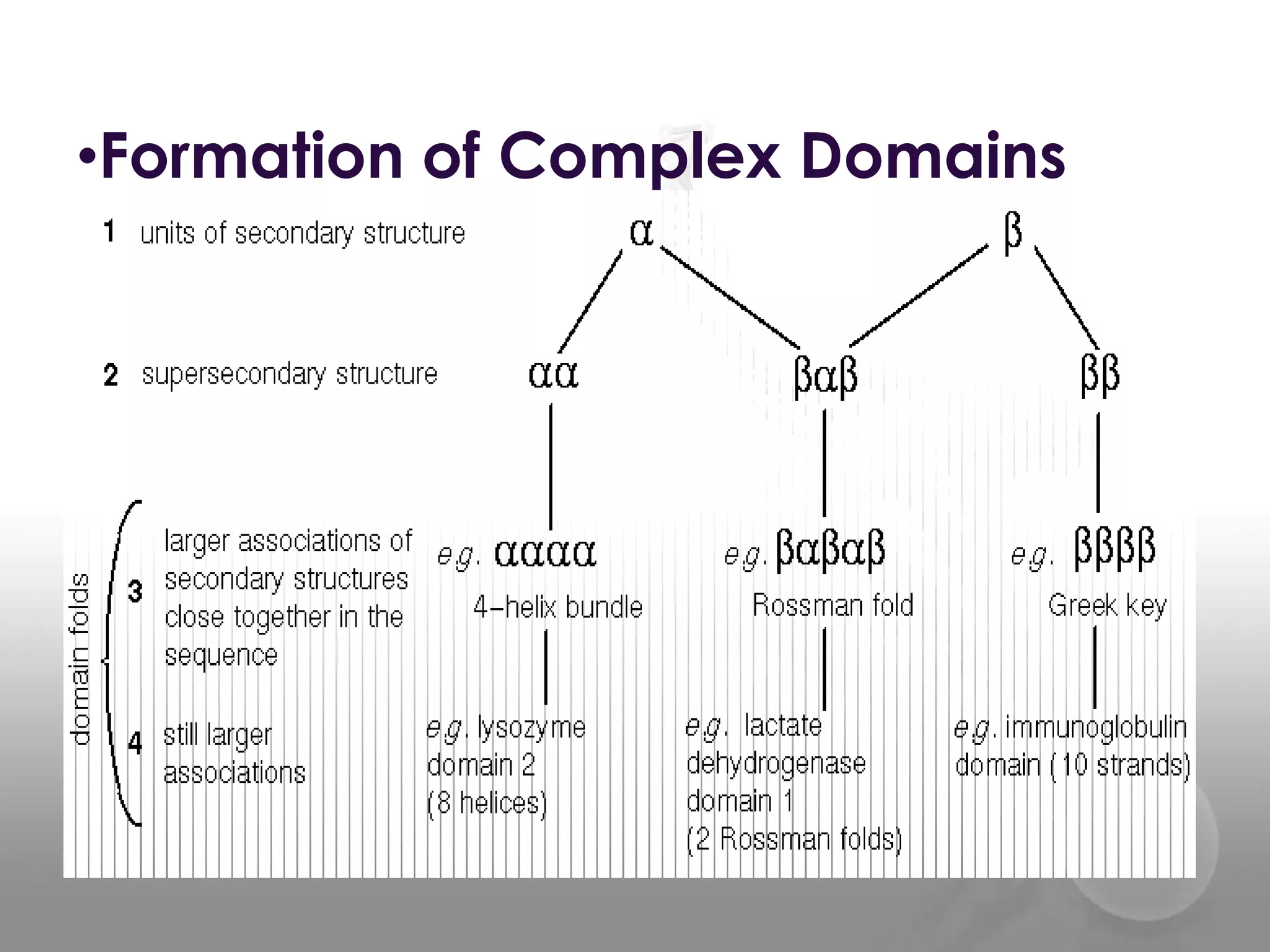
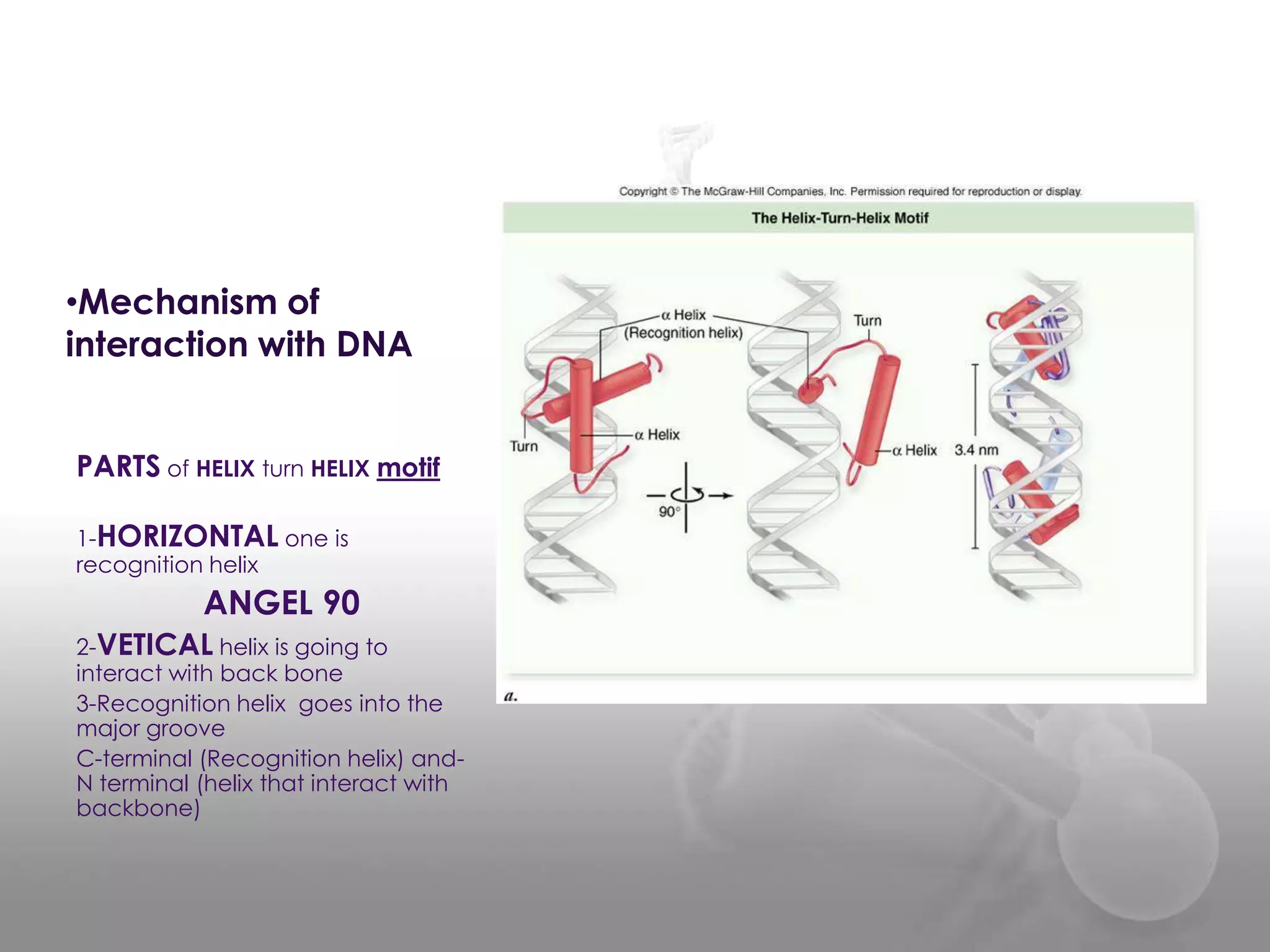
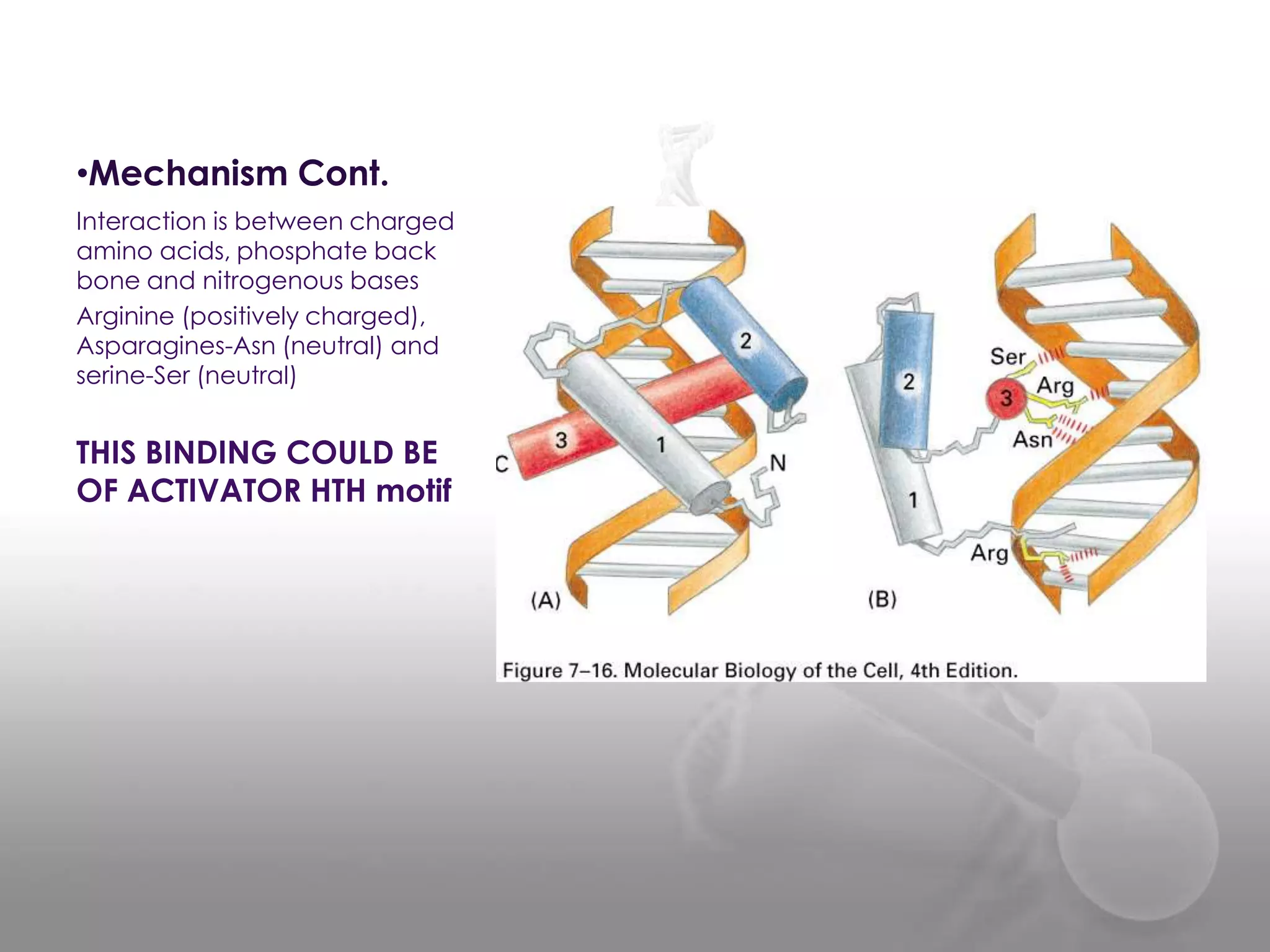
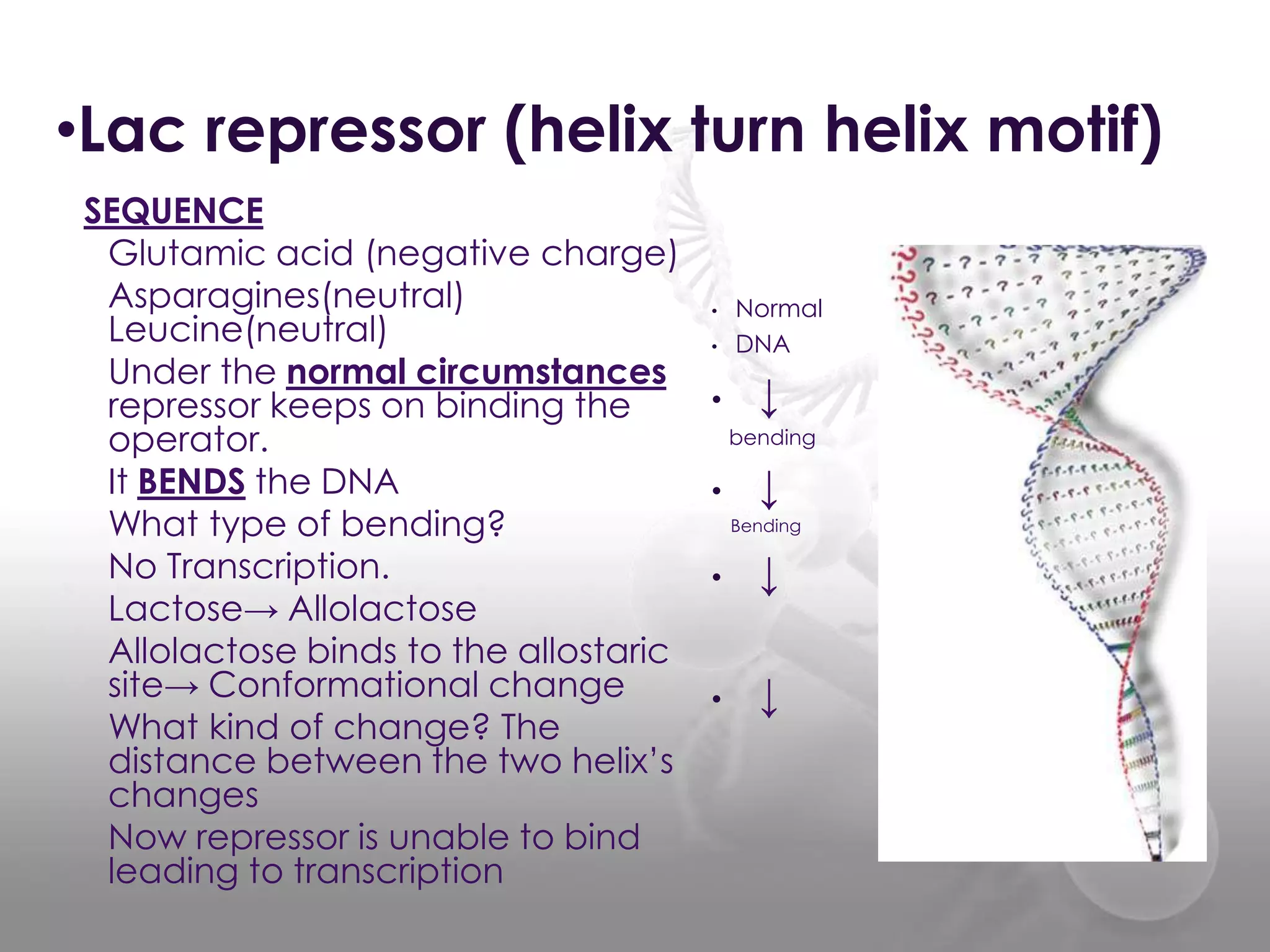
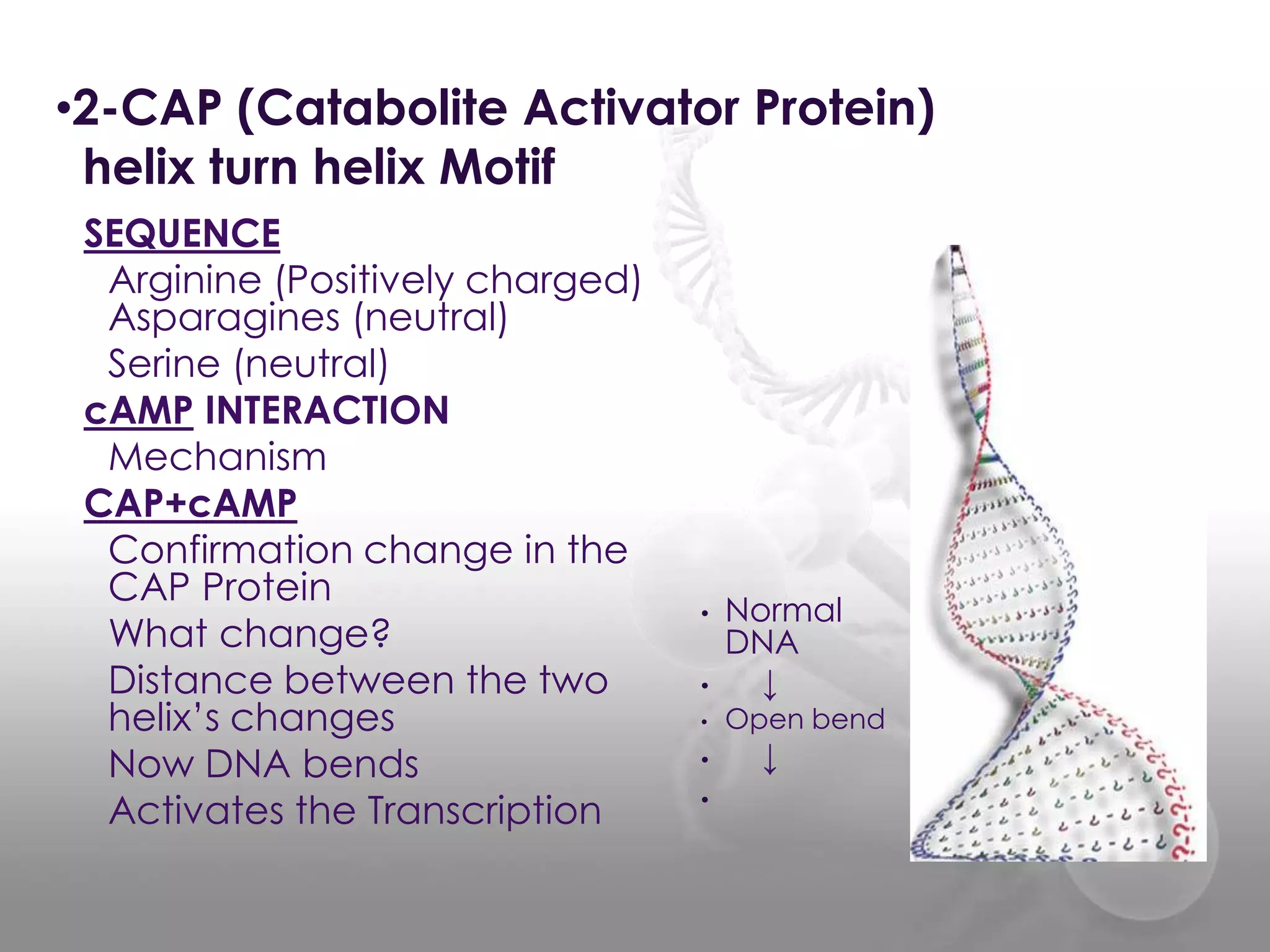
The document discusses the DNA binding domain known as the helix-turn-helix structure. It contains two alpha helices joined by a turn that interacts with DNA in the major groove through charged amino acids on the recognition helix. The helix-turn-helix motif can function in both gene activation and repression by changing its conformation through modifications or allosteric binding, thereby changing the DNA bending and accessibility of transcriptional machinery. Examples given are the lac repressor, which bends DNA and prevents transcription until binding lactose, and the catabolite activator protein (CAP), which activates transcription after cAMP binding causes a conformational change in the helix-turn-helix motif.