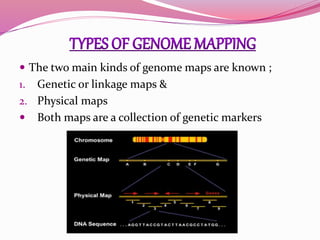
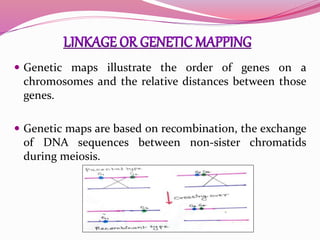
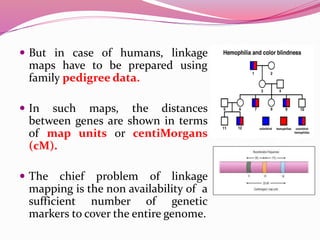
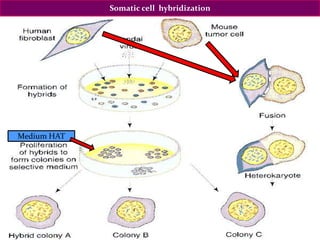
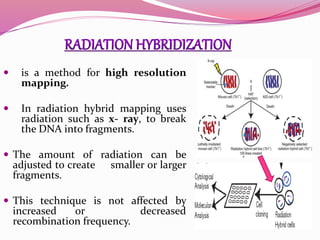
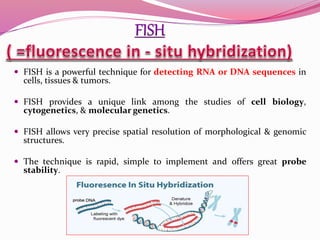
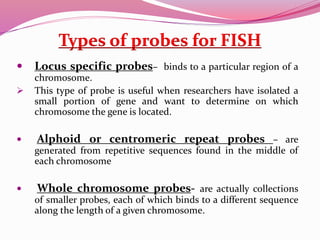
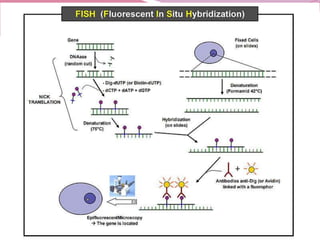
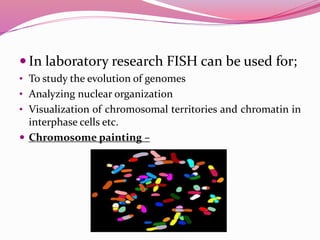
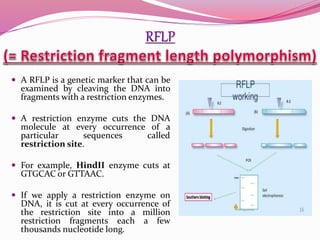
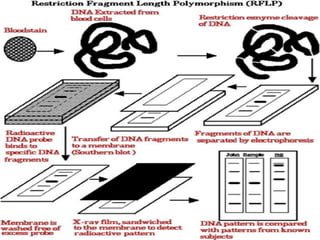
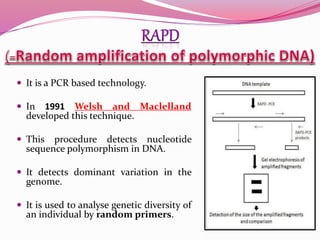
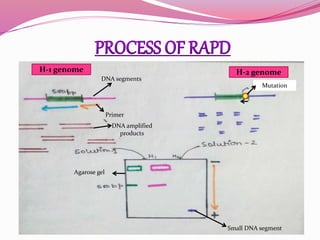
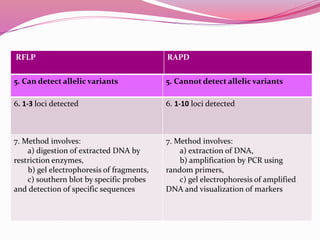
The document discusses genome mapping, detailing its significance in genetics by illustrating the structure and arrangement of genes within chromosomes. It outlines the two main types of genome maps—genetic and physical maps—along with techniques such as somatic cell hybridization and fluorescent in situ hybridization (FISH) that are used for gene mapping and detecting DNA sequences. Additionally, it compares methods like RFLP and RAPD for analyzing genetic diversity, their applications in various fields, and highlights the challenges and benefits associated with these techniques.