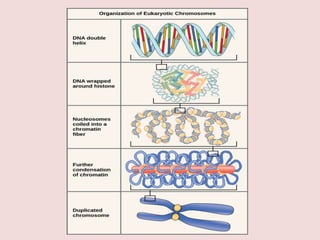
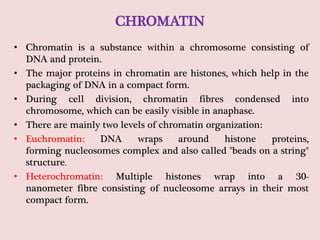
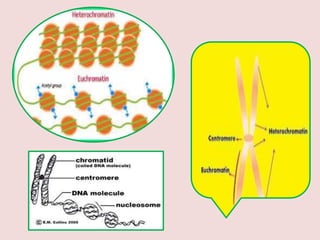
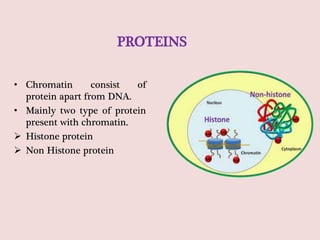
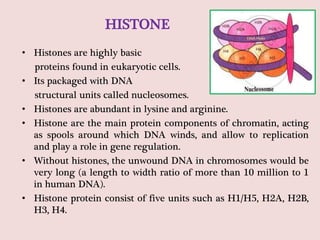
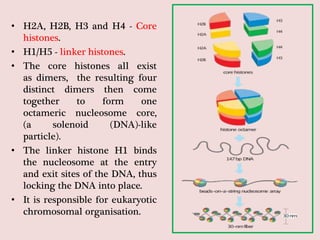
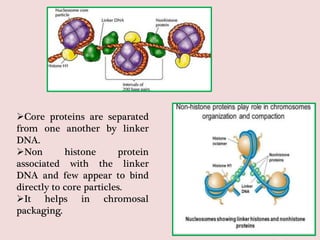
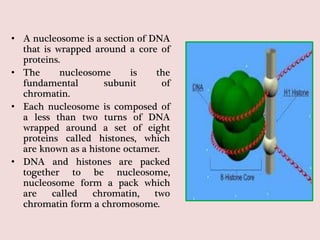
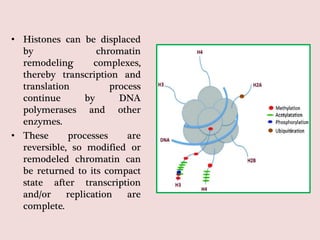
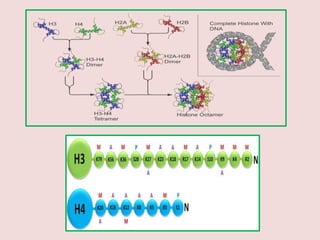
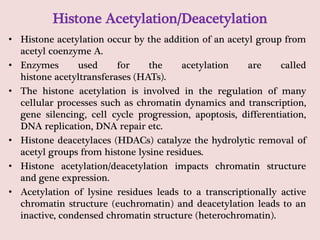
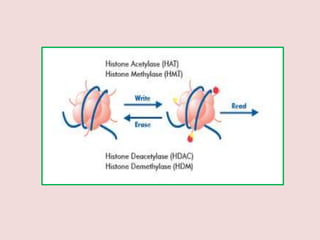
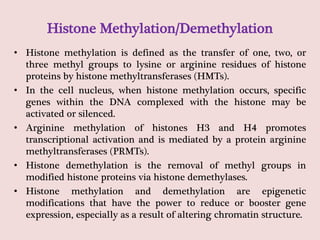
The document summarizes eukaryotic chromosomal organization. It discusses that eukaryotic chromosomes contain linear DNA molecules that are highly compacted through wrapping around histone proteins. This forms chromatin, which exists in two forms - euchromatin that is loosely packed and facilitates transcription, and heterochromatin that is tightly packed and prevents transcription. The document outlines the various levels of compaction from DNA to nucleosomes to chromatin fibers and chromosomes, and describes the roles and modifications of histone and non-histone proteins in chromatin organization and gene regulation.