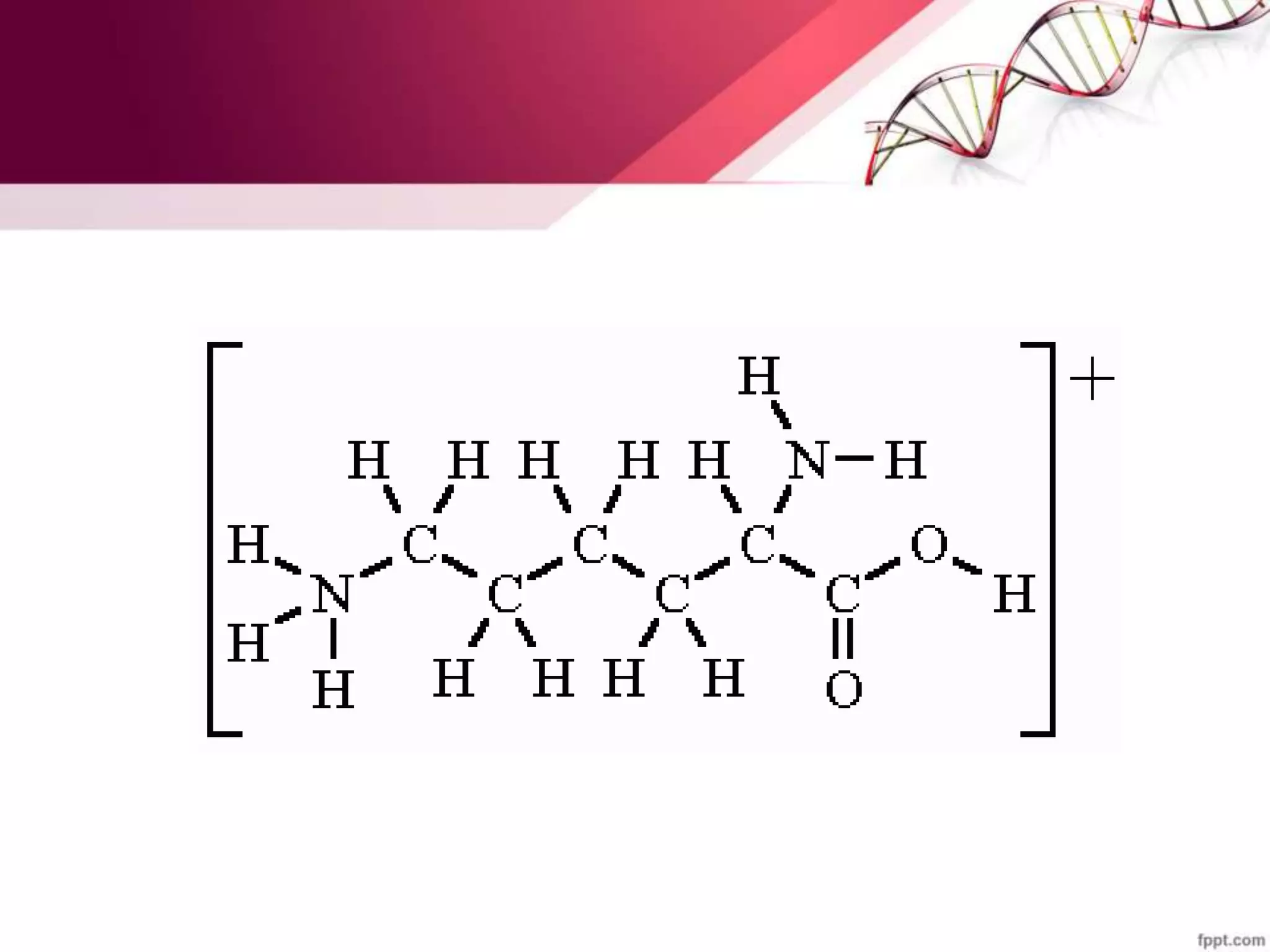
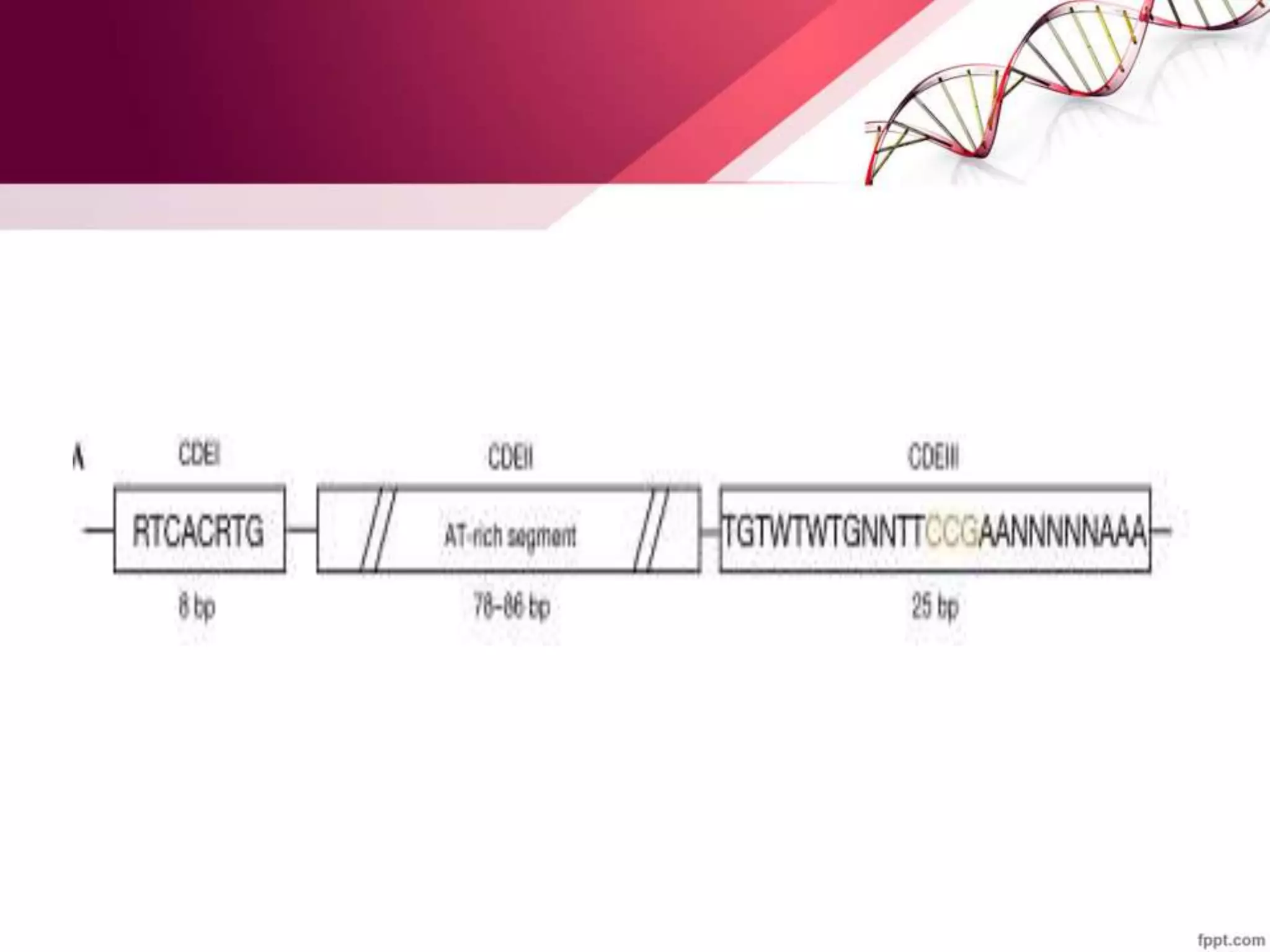
The document summarizes yeast chromosome structure and function. It discusses how DNA is packaged into chromatin and nucleosomes in yeast. The yeast genome contains 16 chromosomes with centromeres and telomeres. Histones can be post-translationally modified through acetylation, deacetylation, and methylation, which impacts chromatin structure and gene expression. Centromeres, origins of replication, and telomeres are also described.