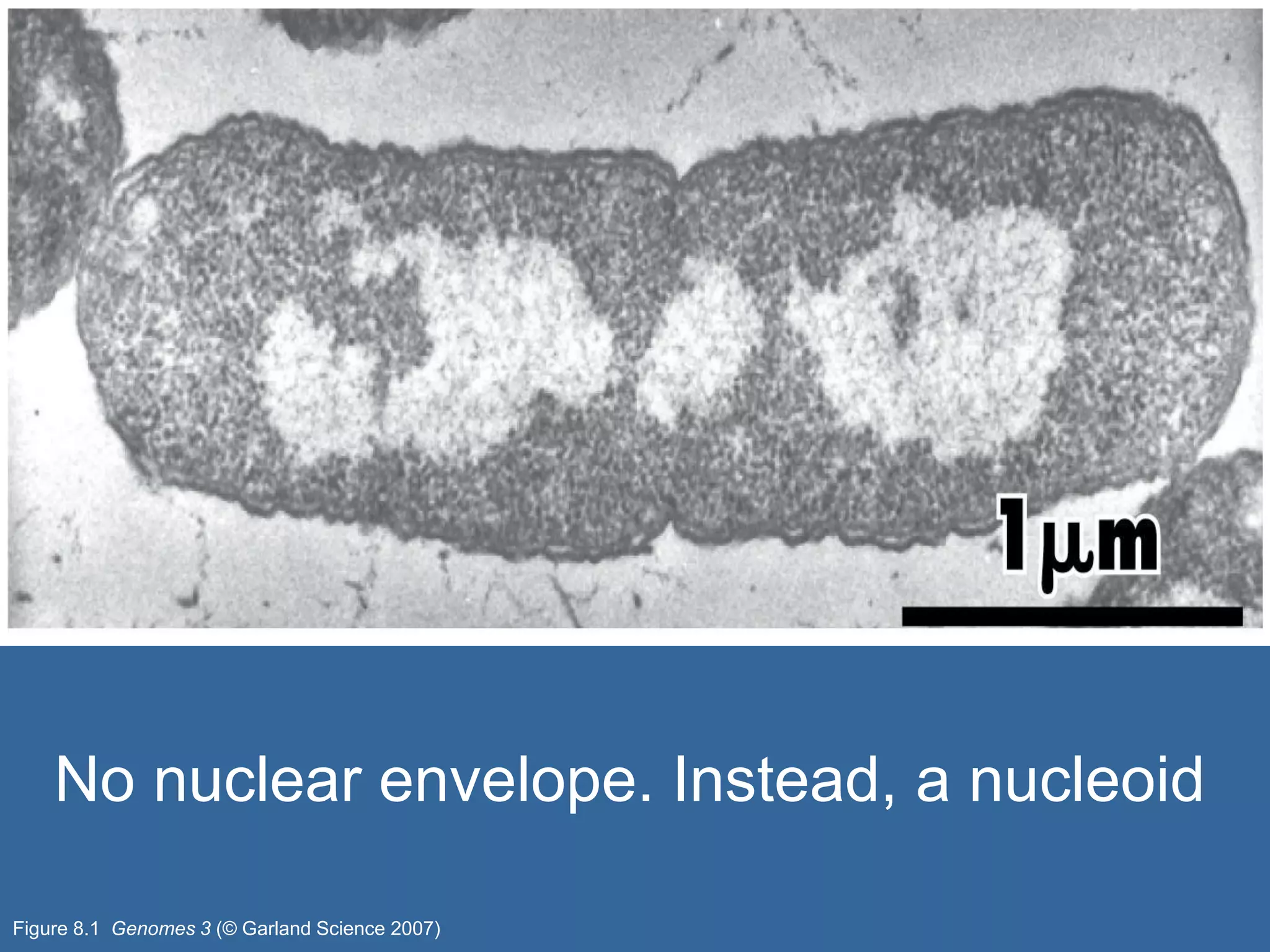
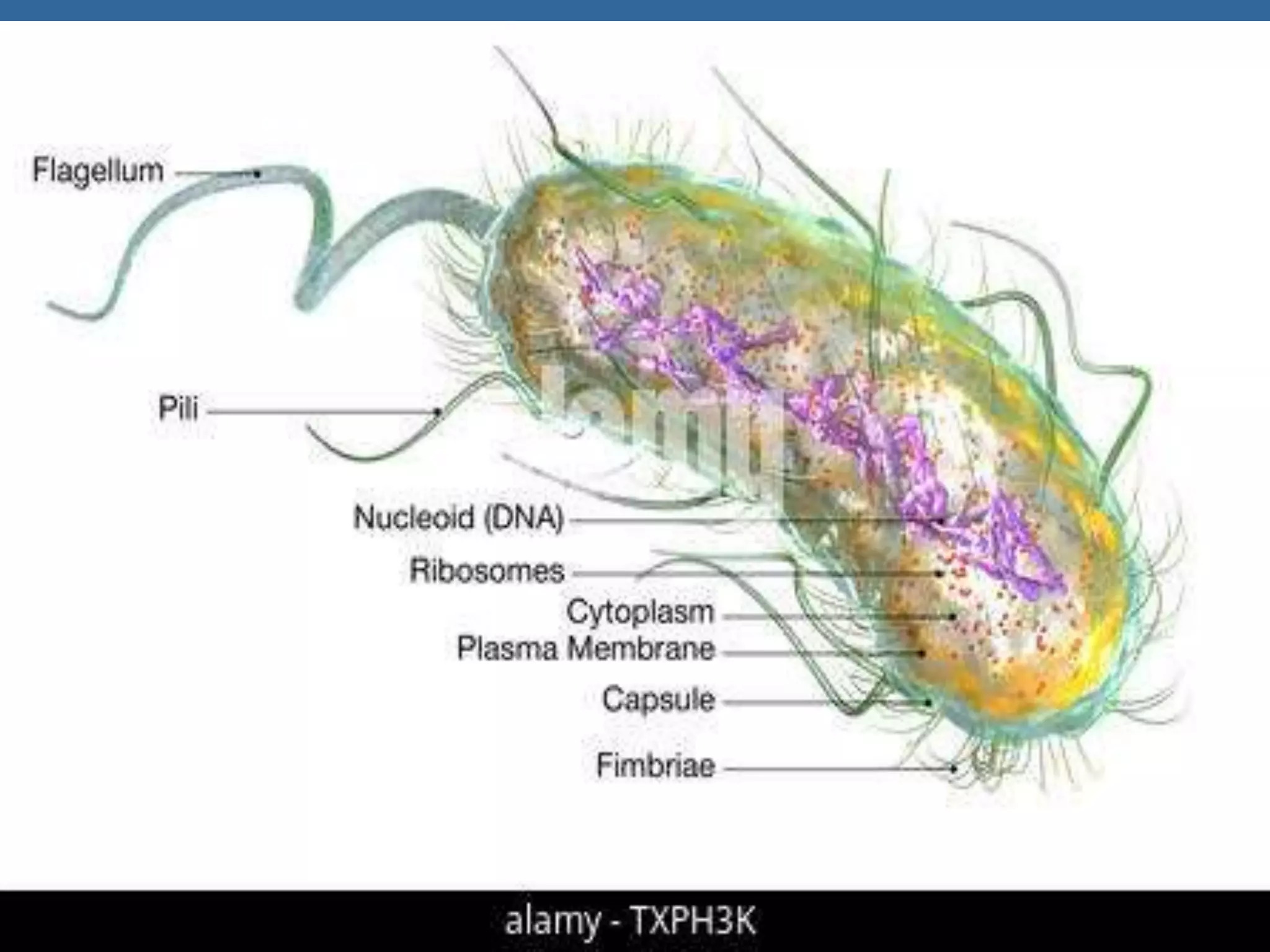
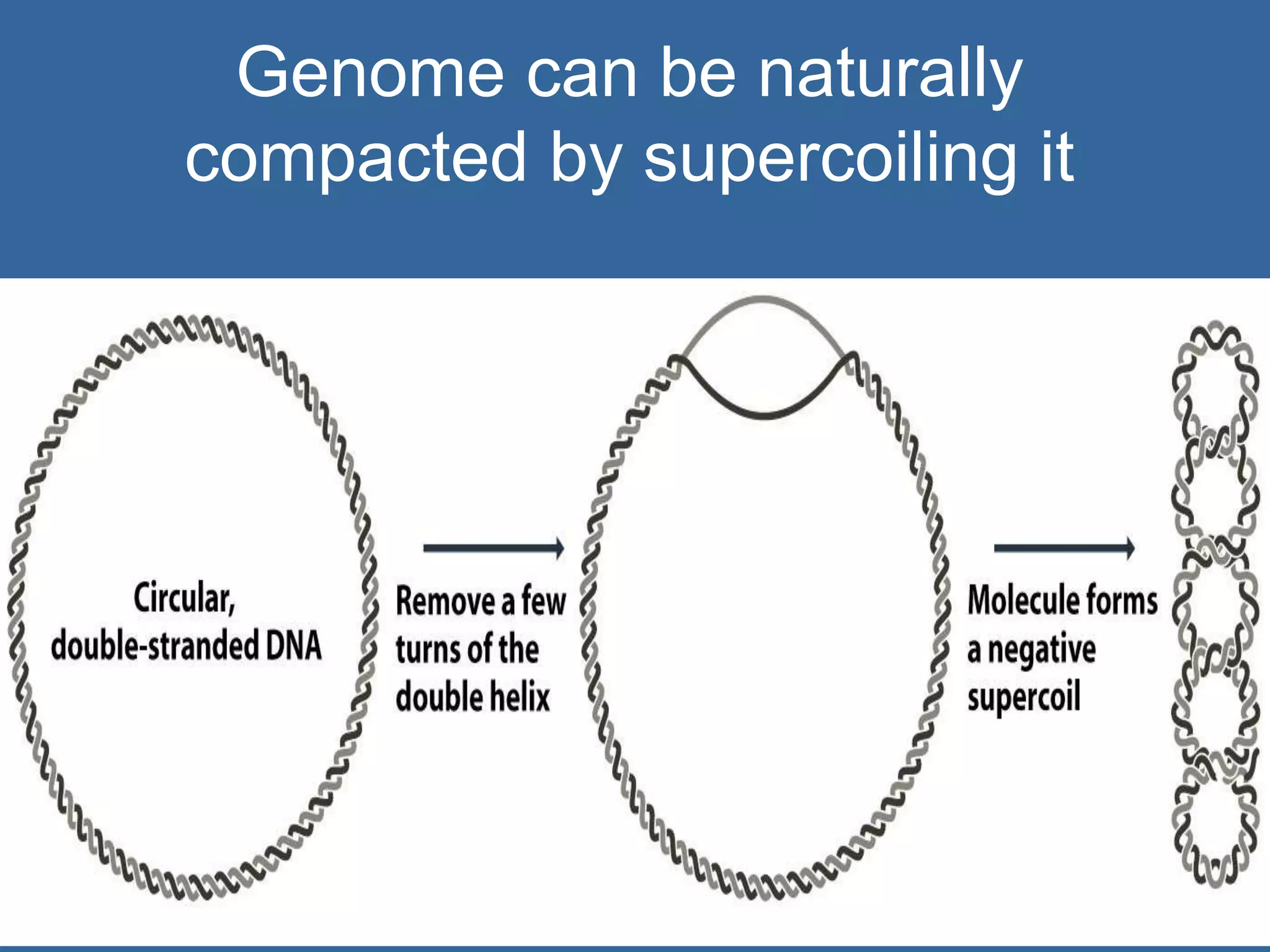
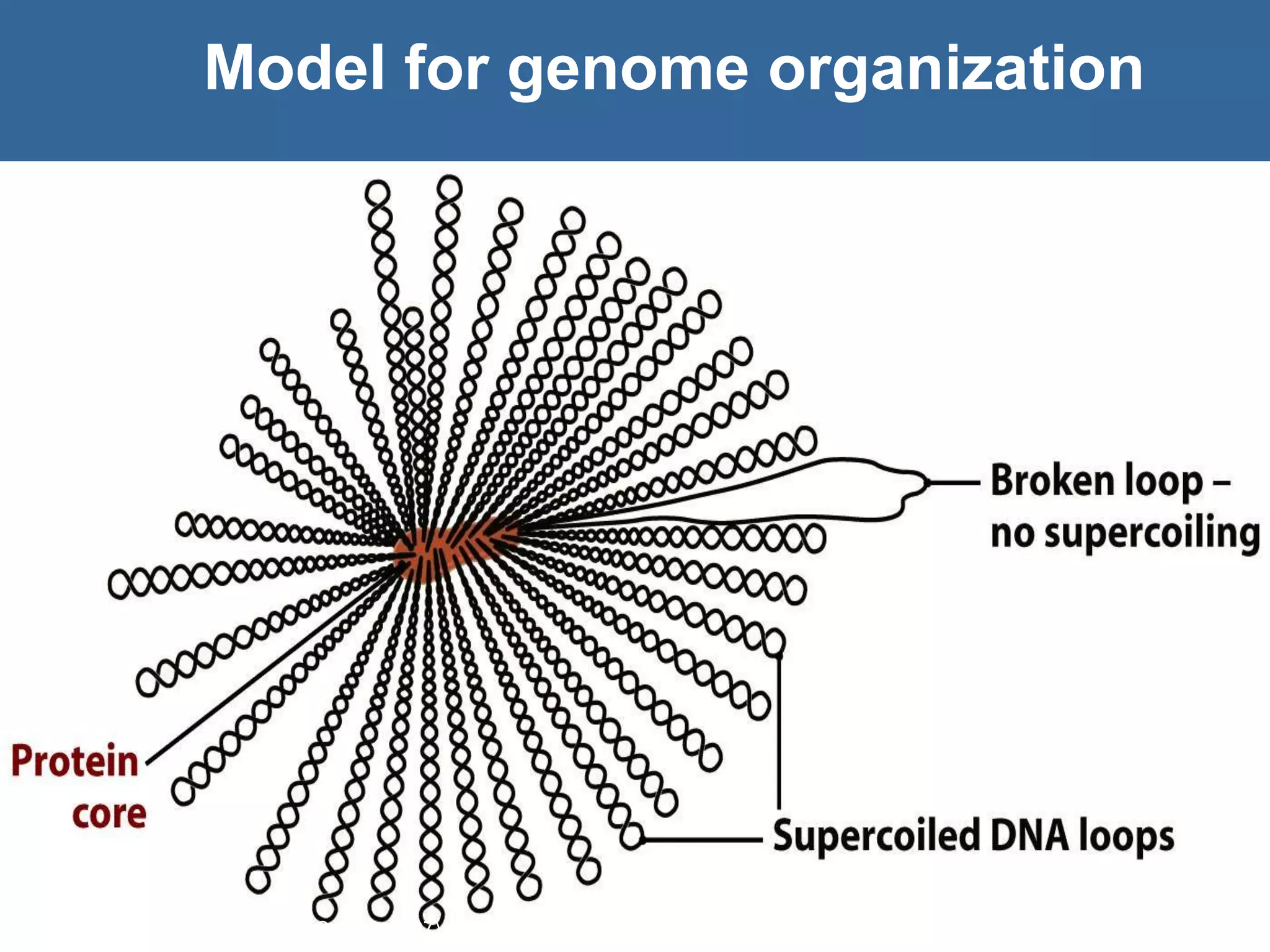
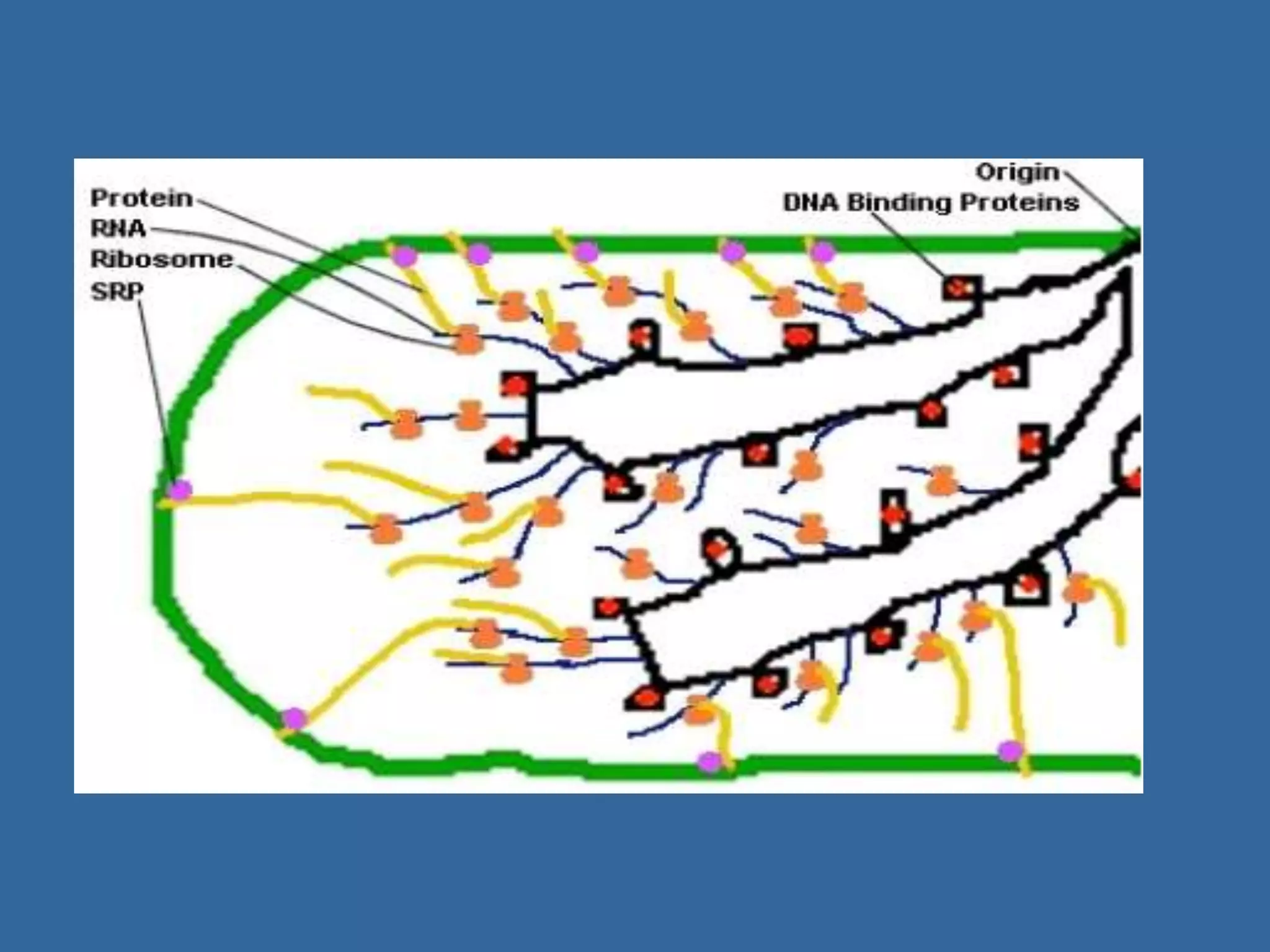
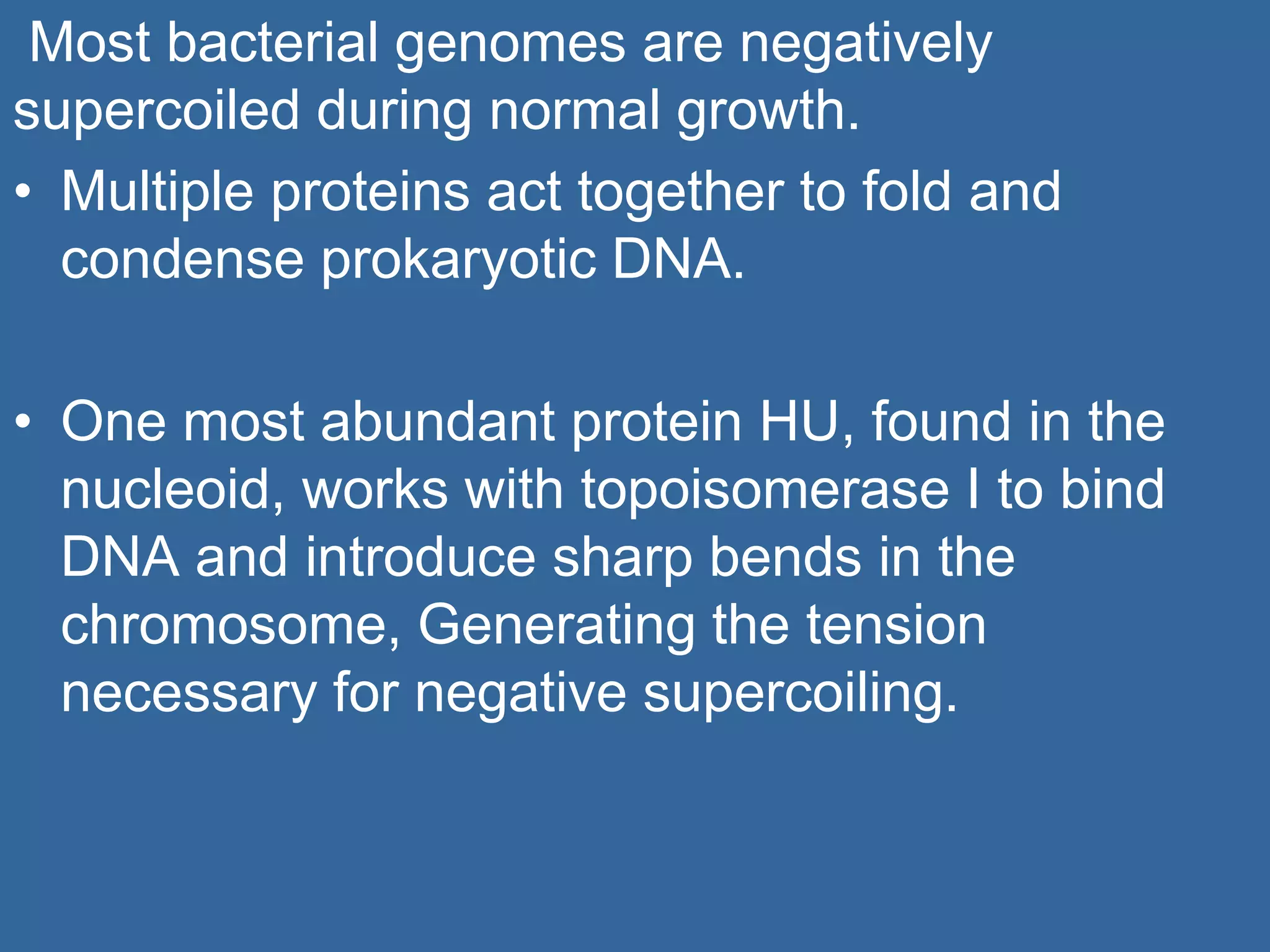
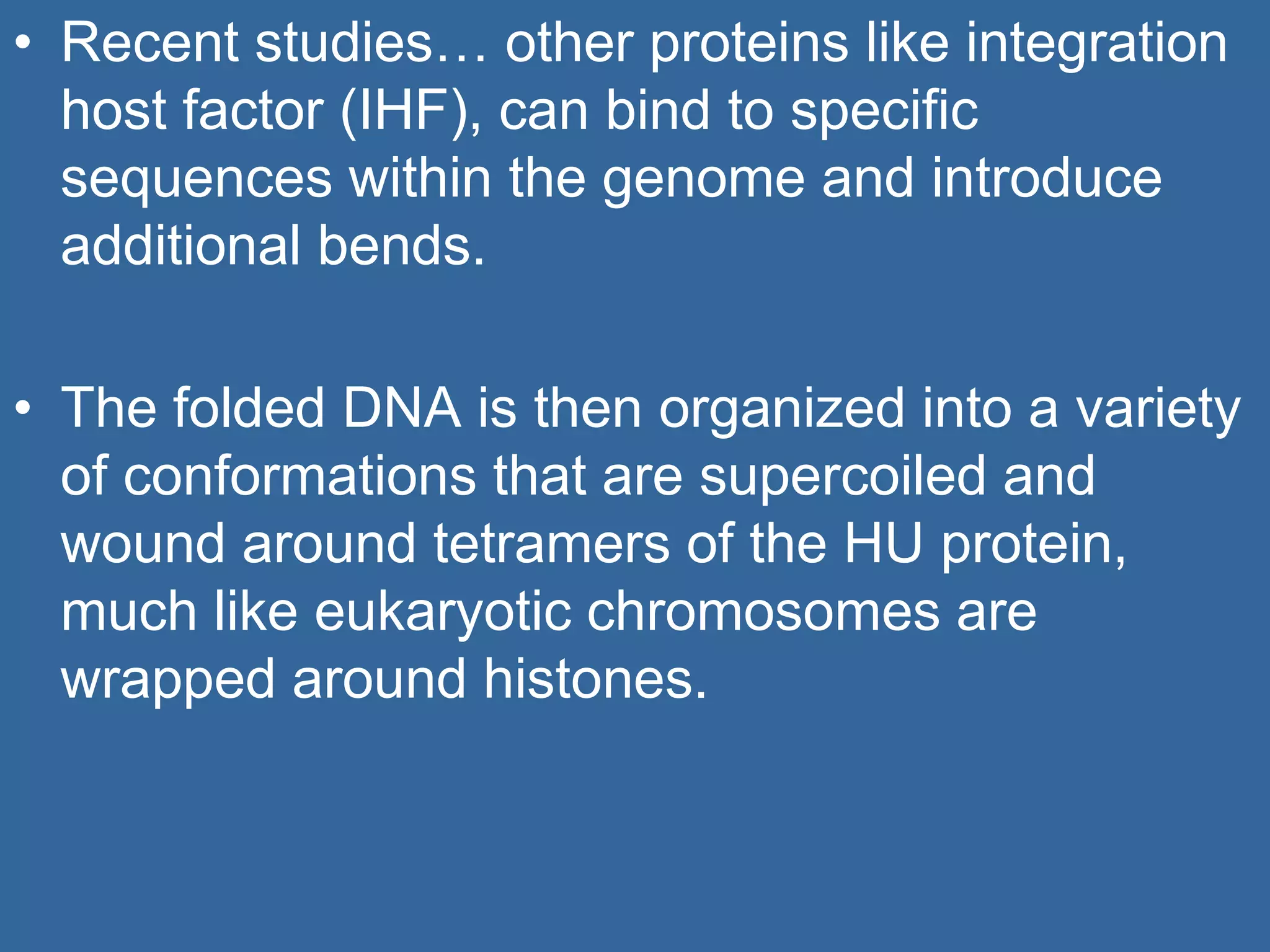
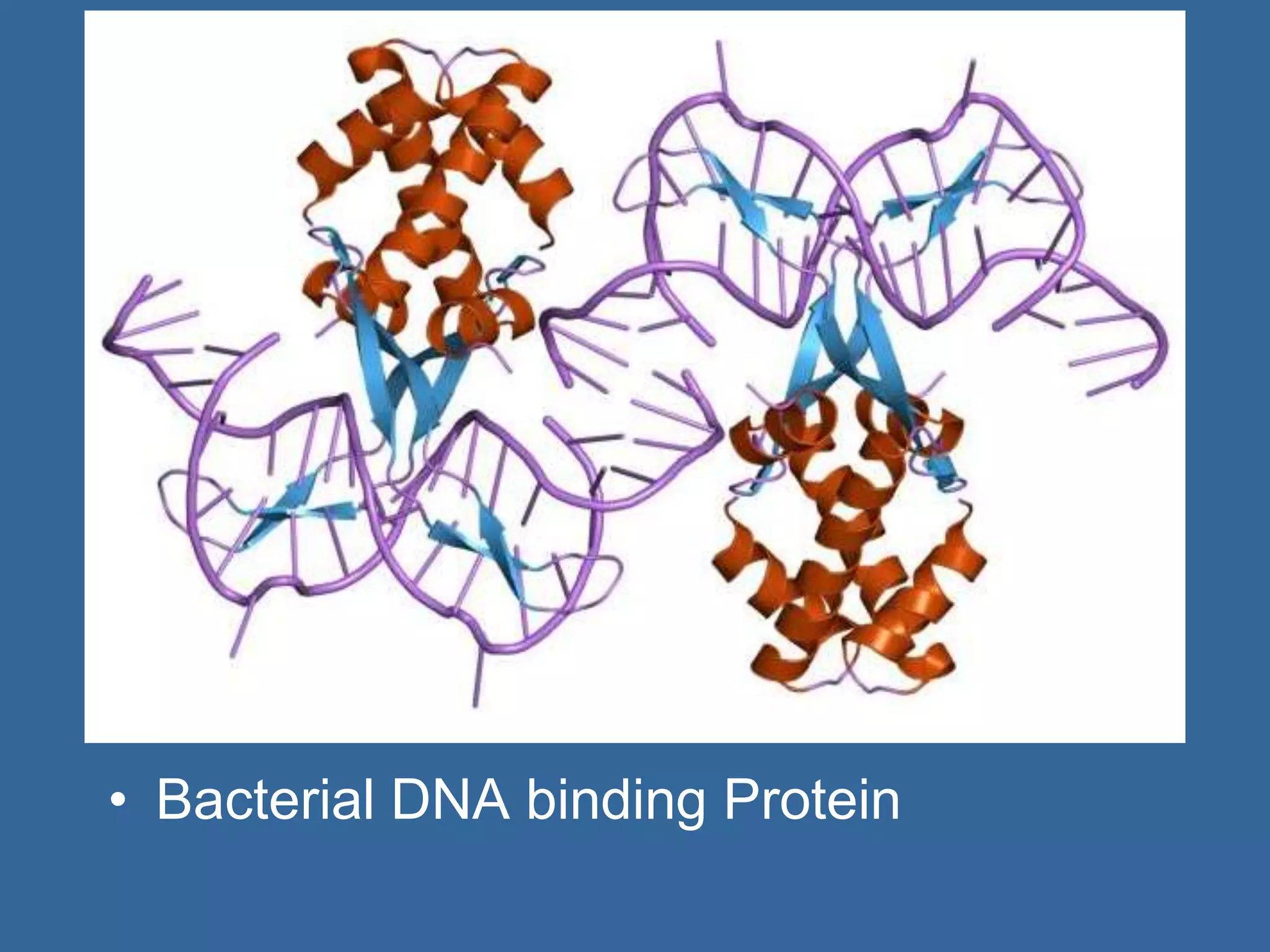
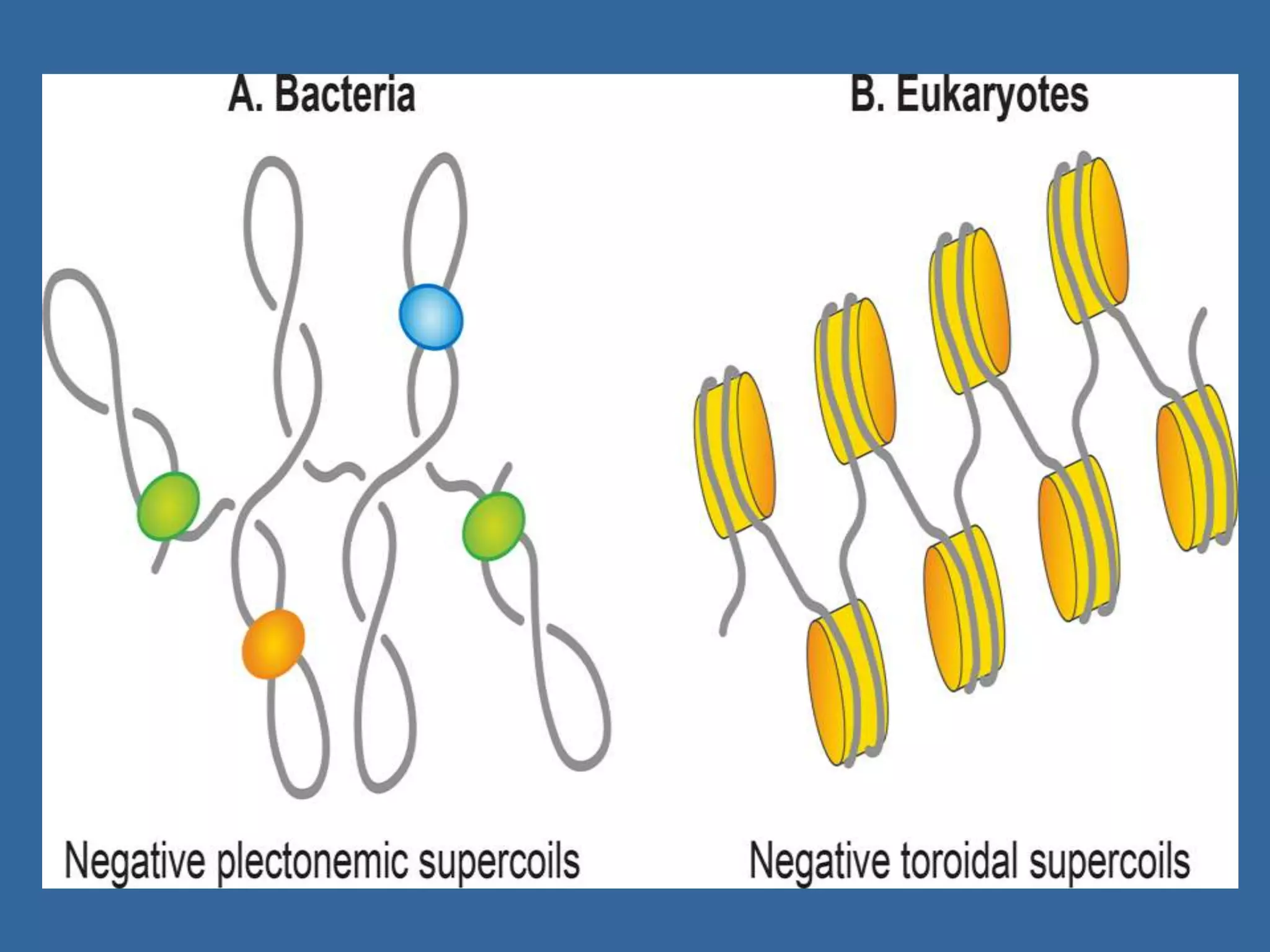
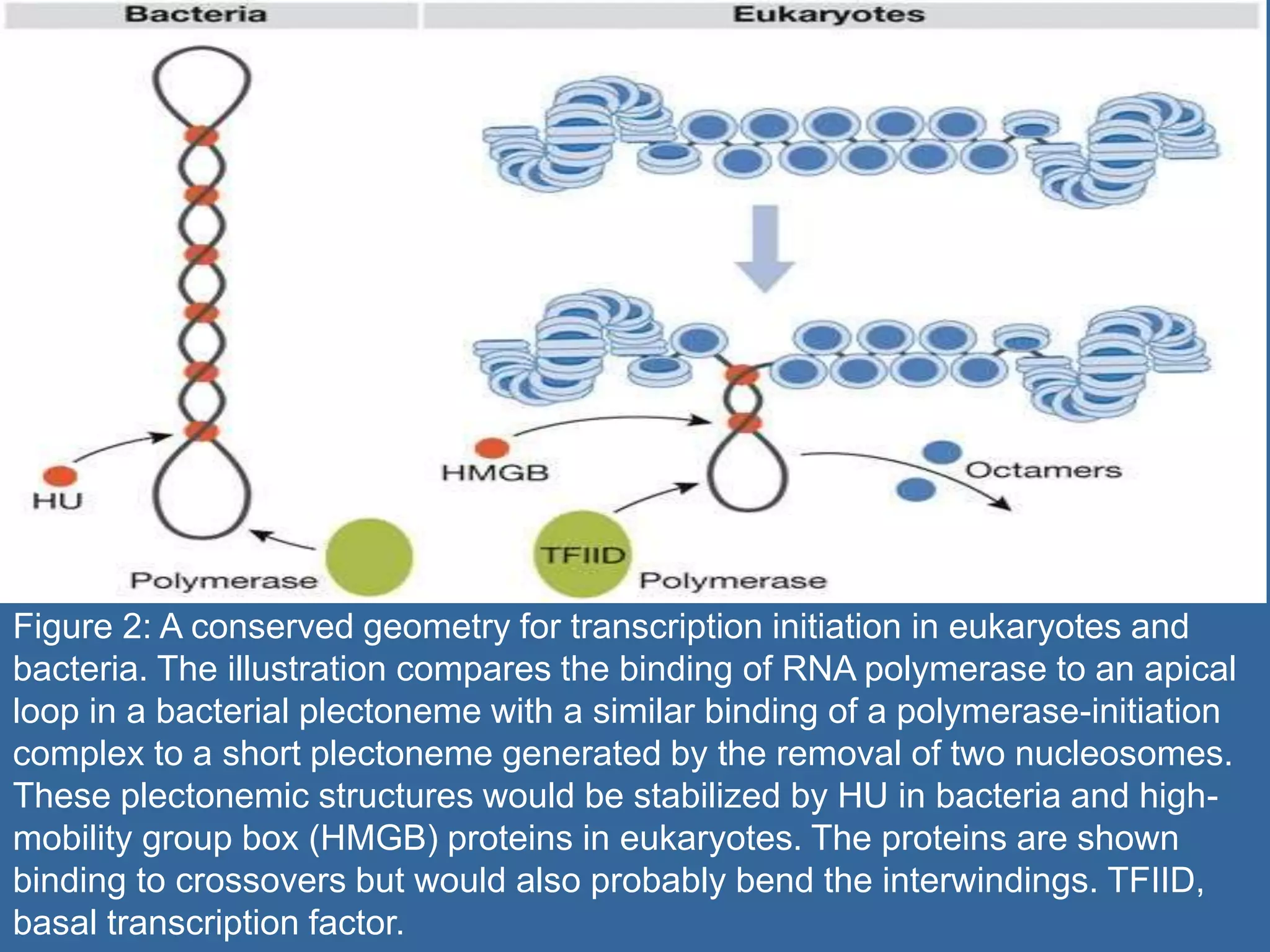
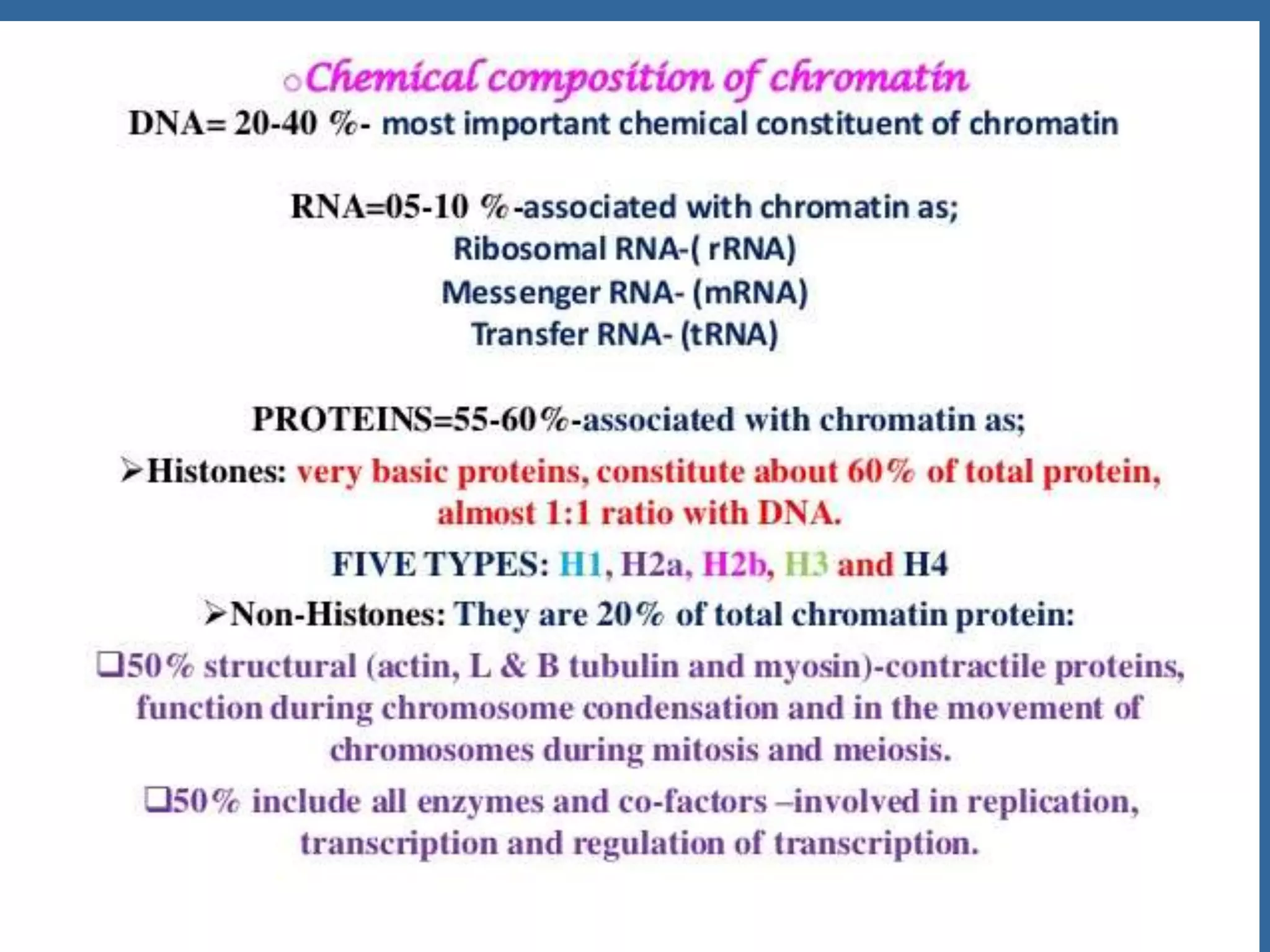
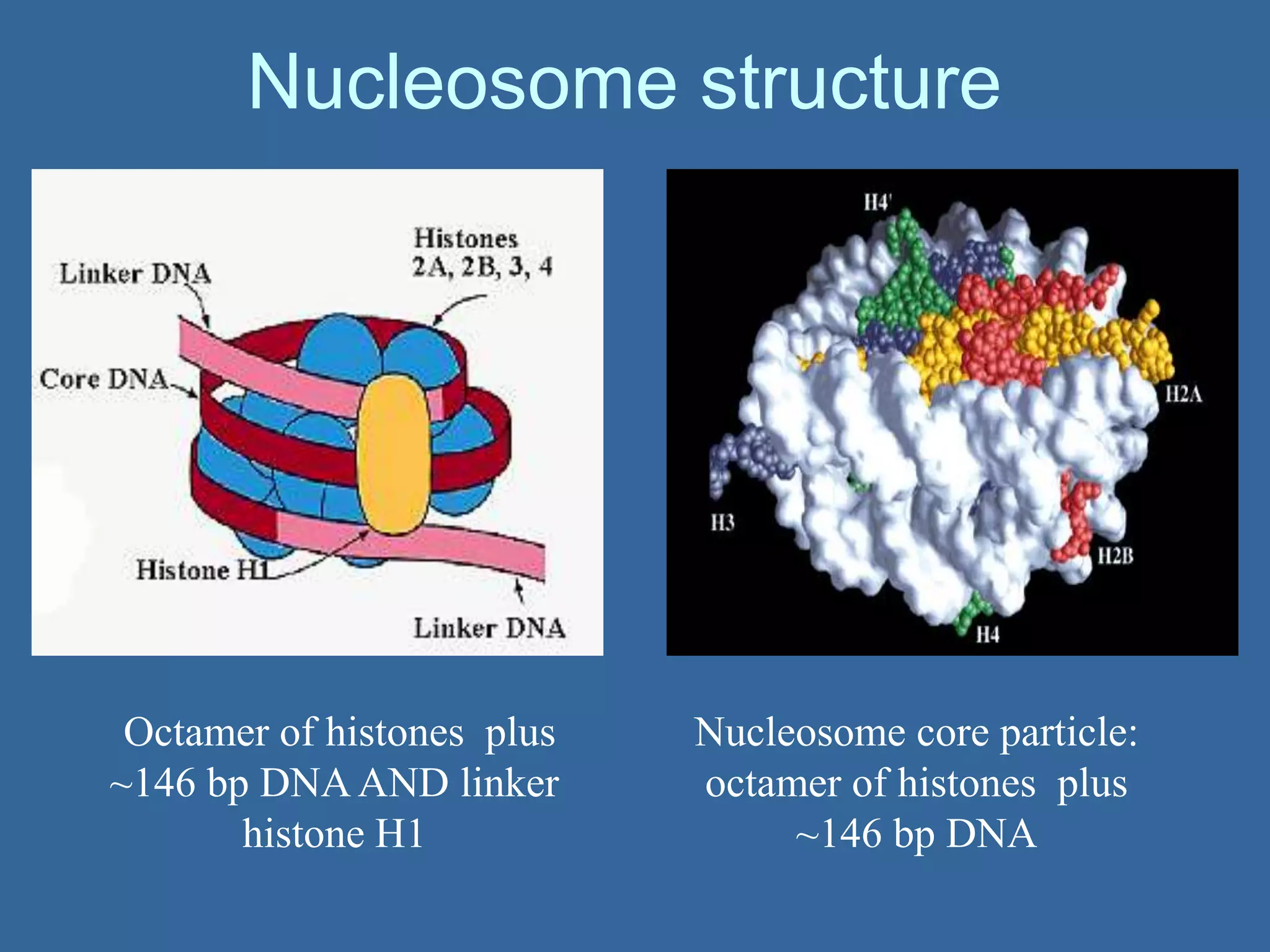
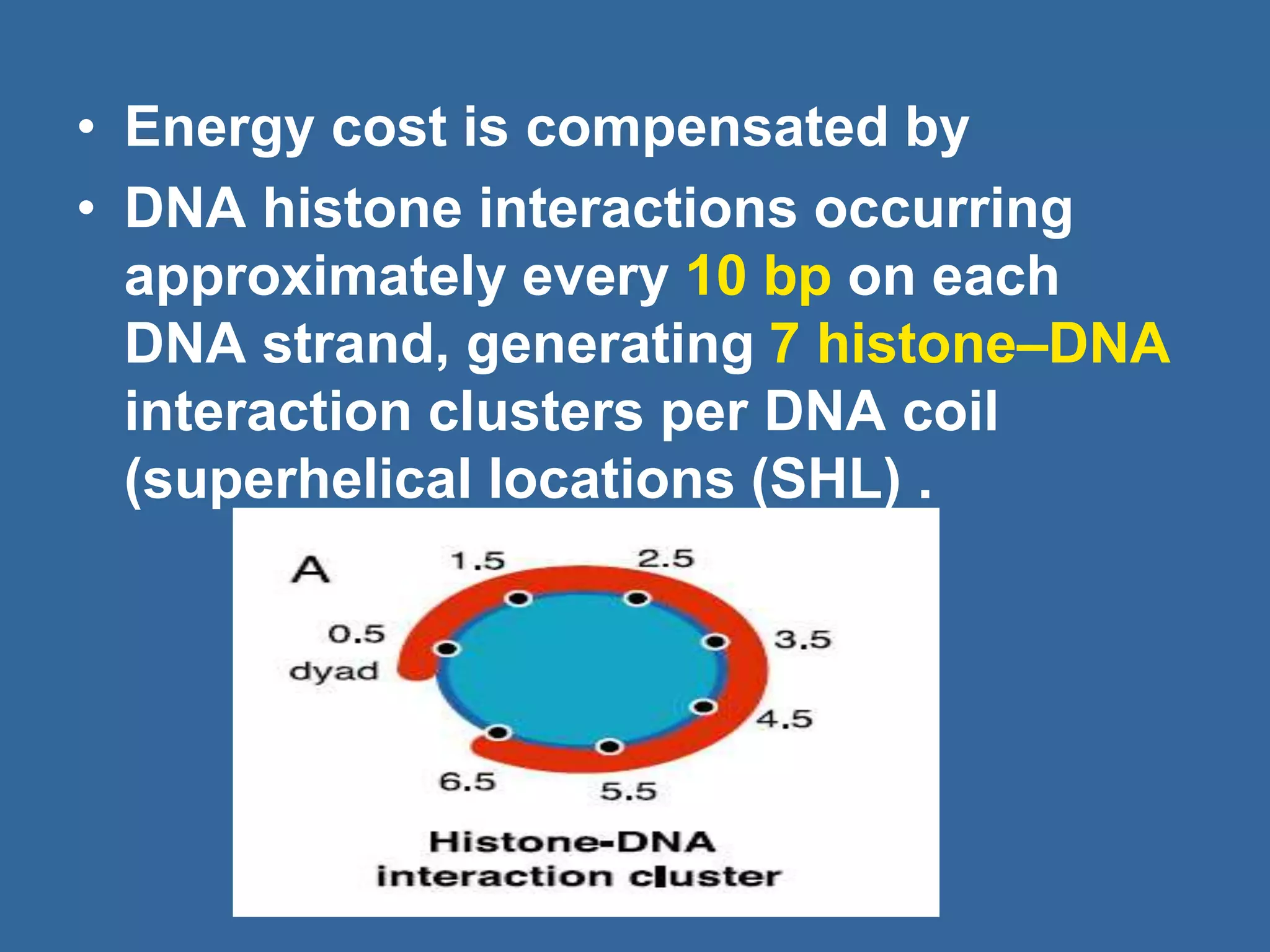
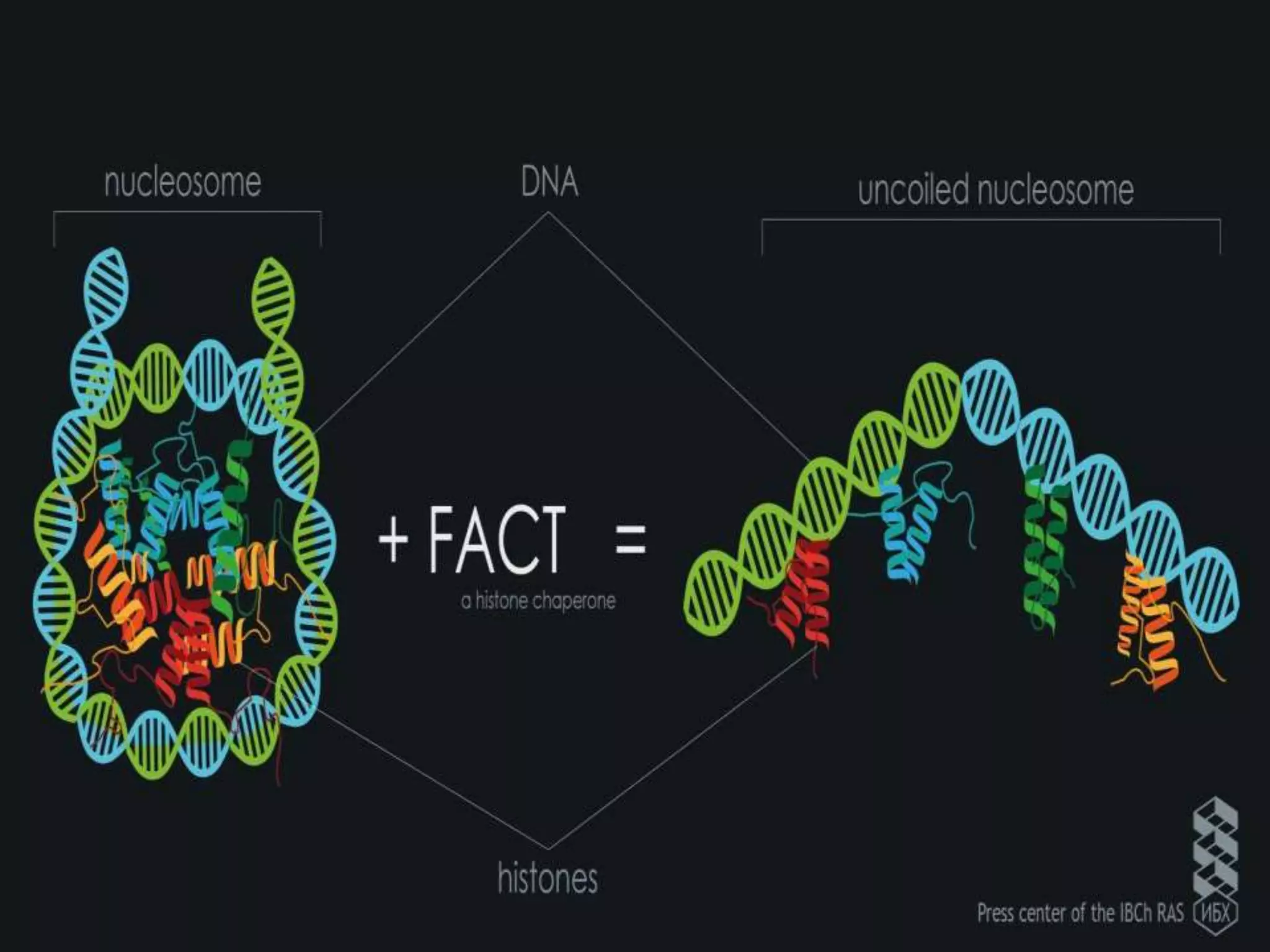
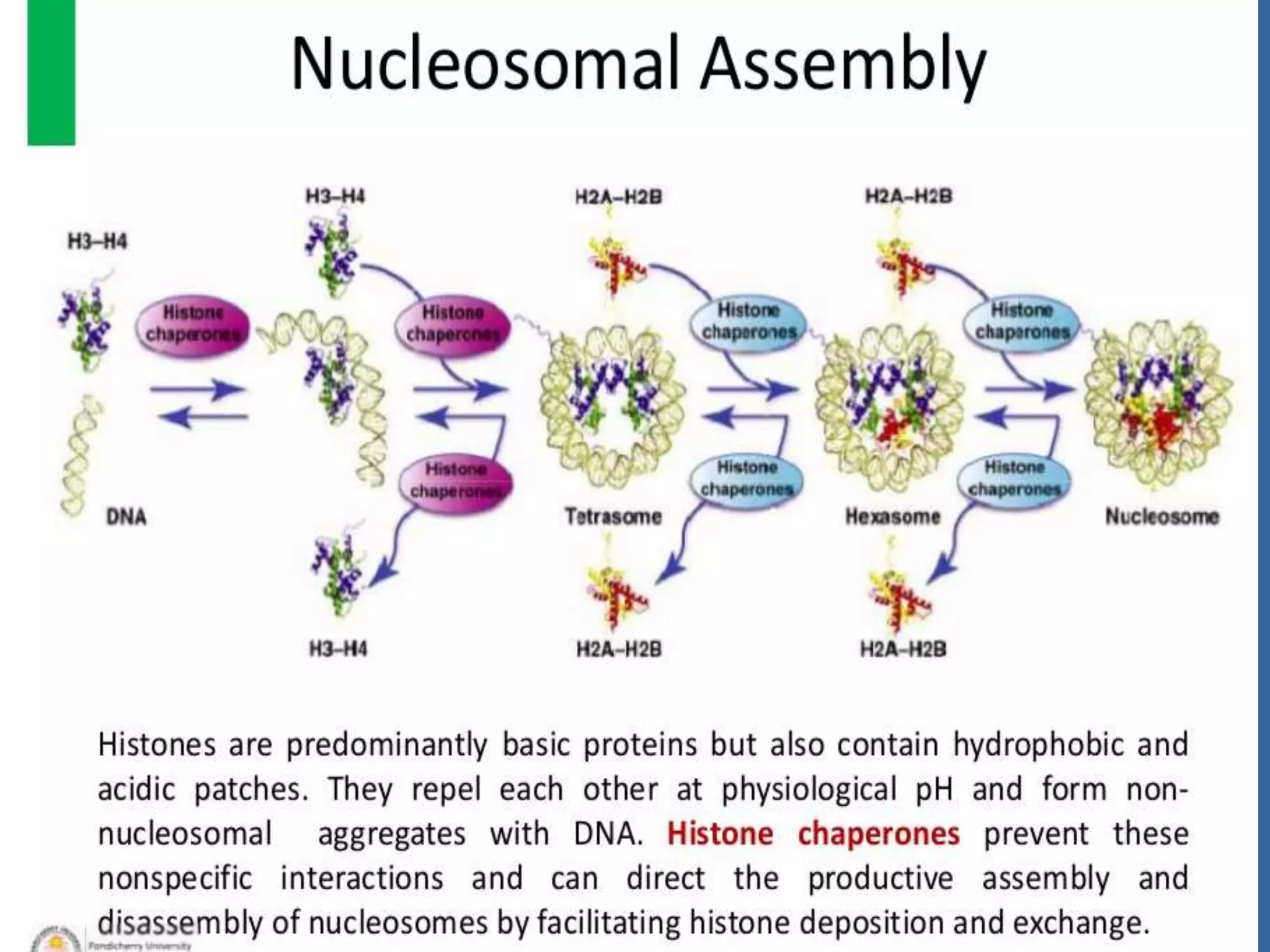
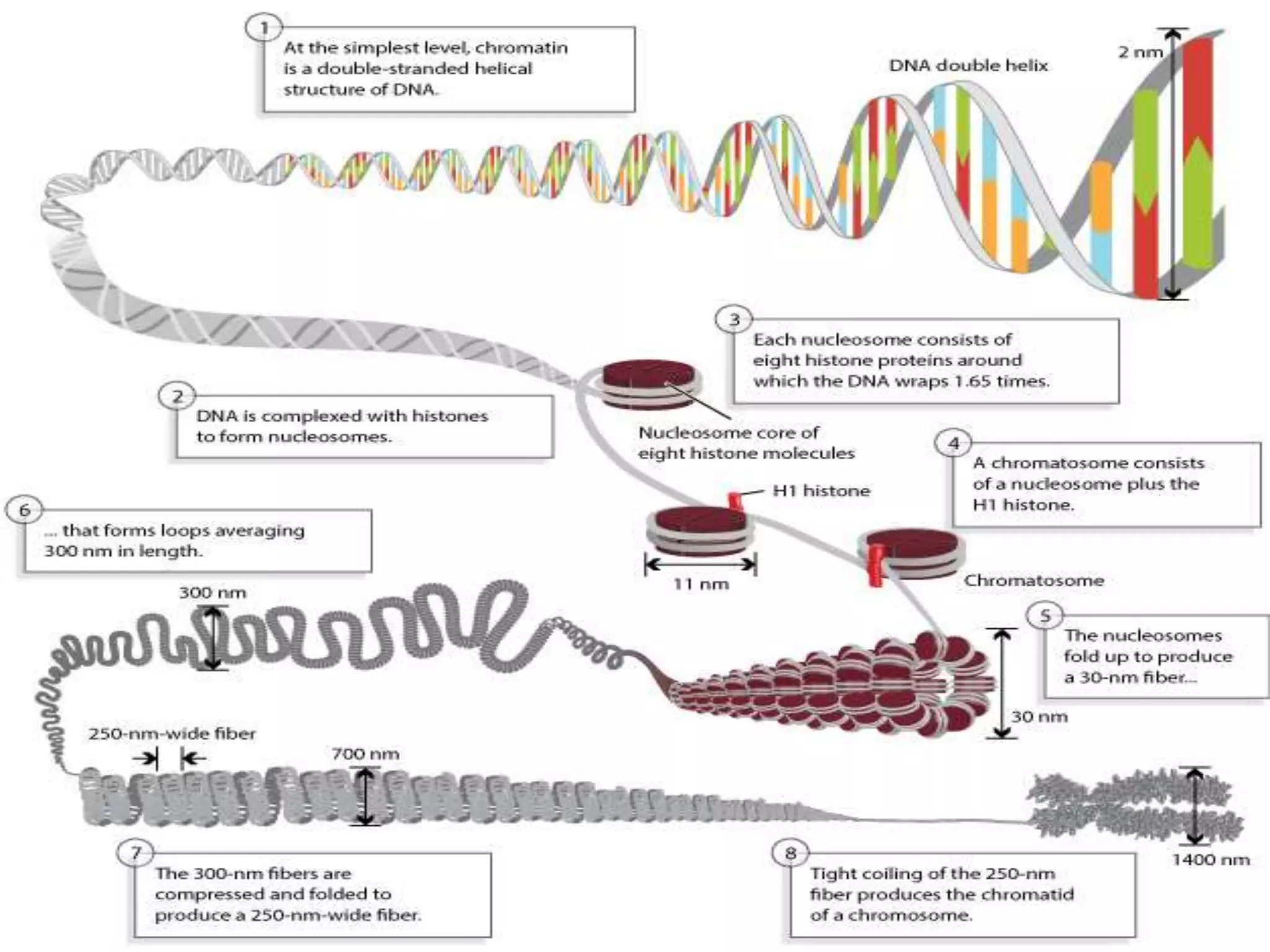
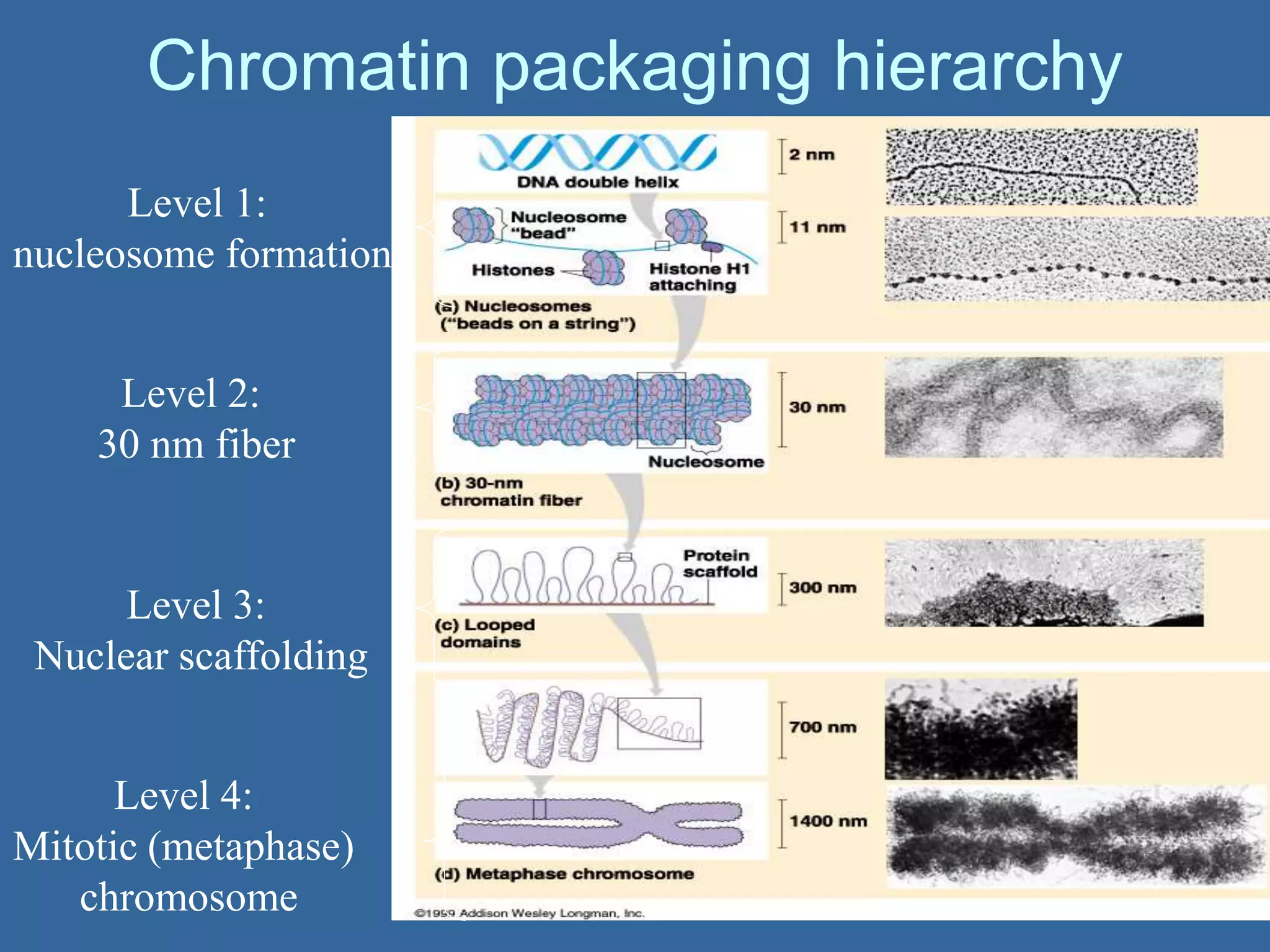
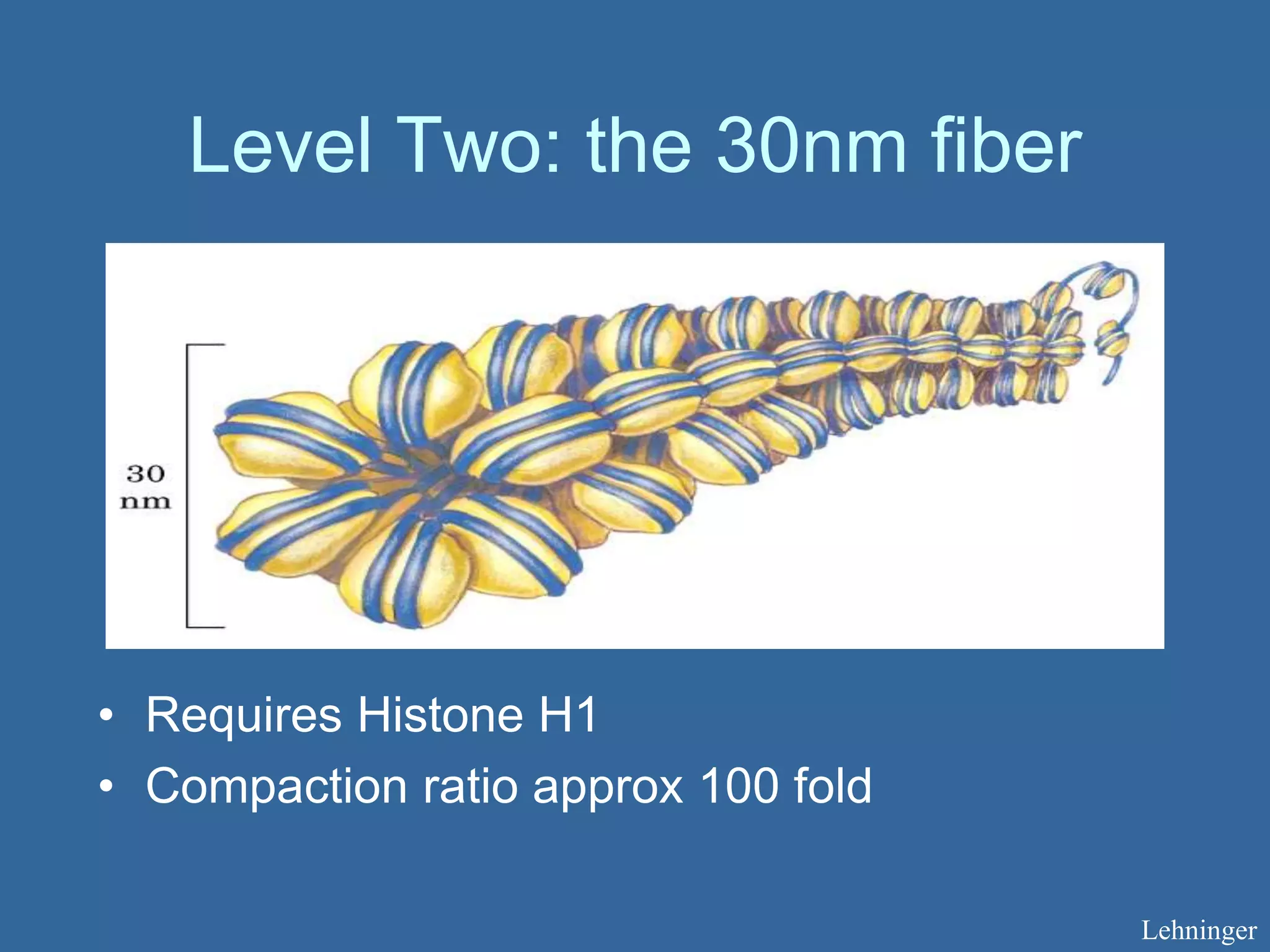
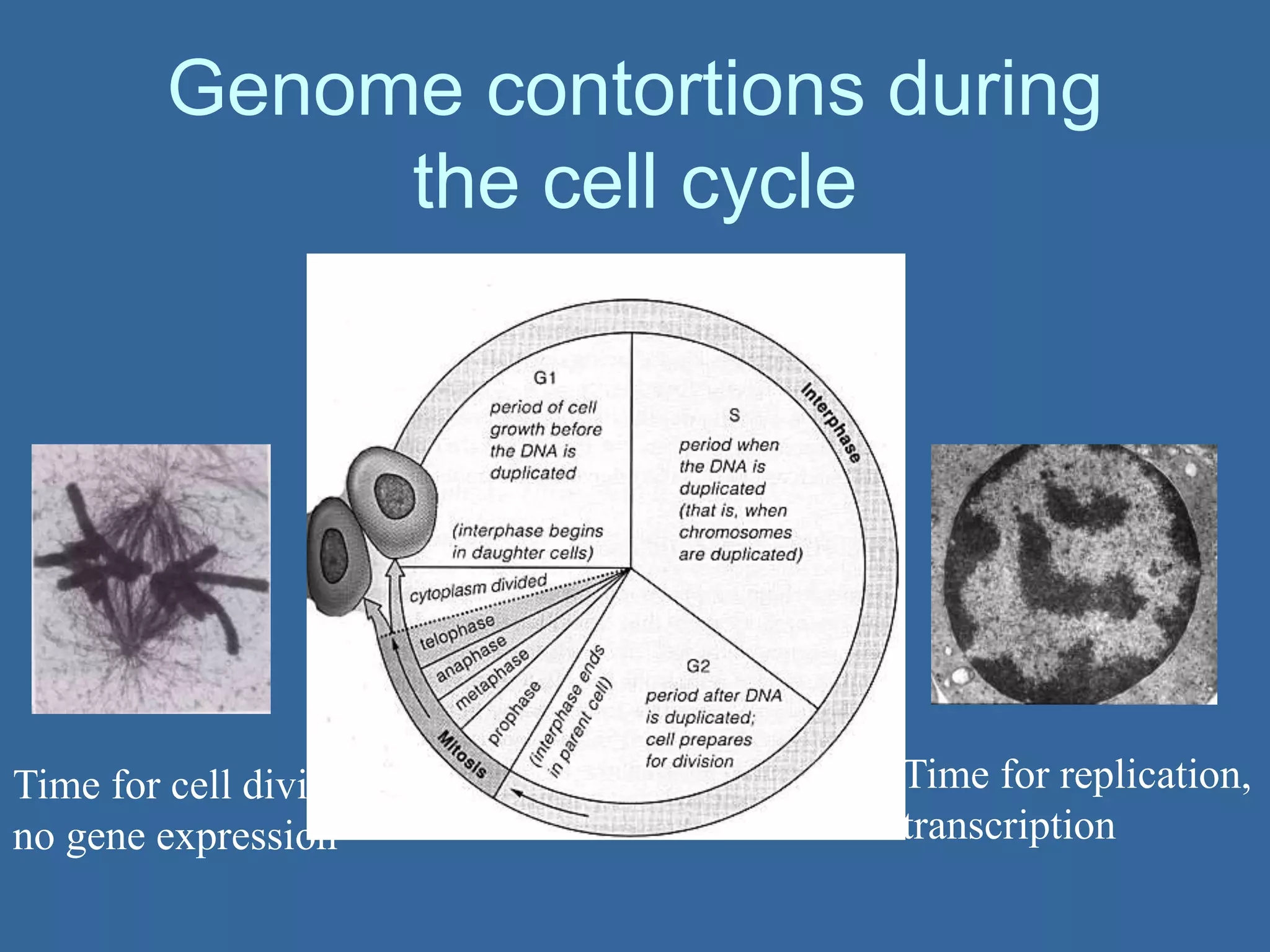
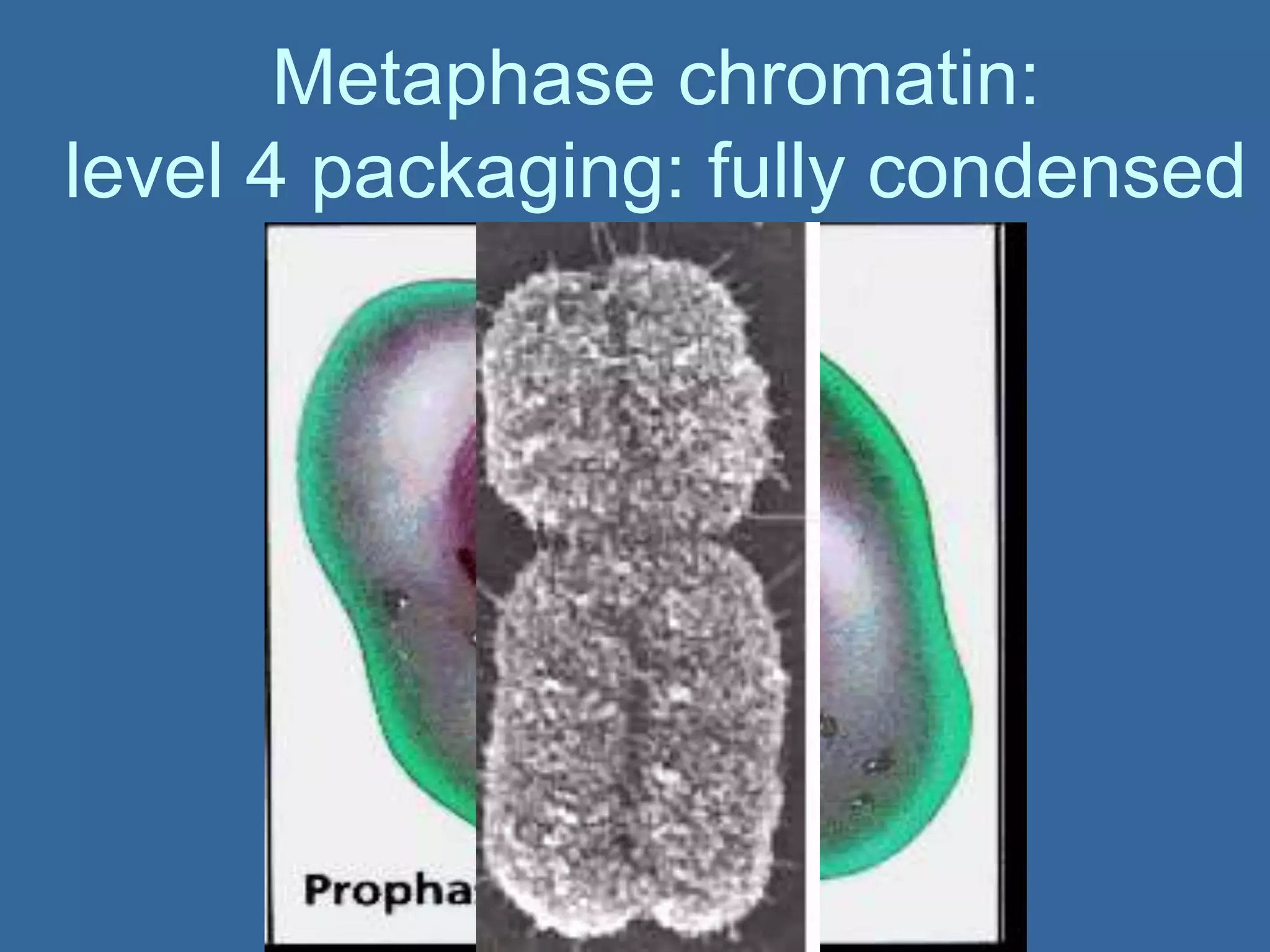
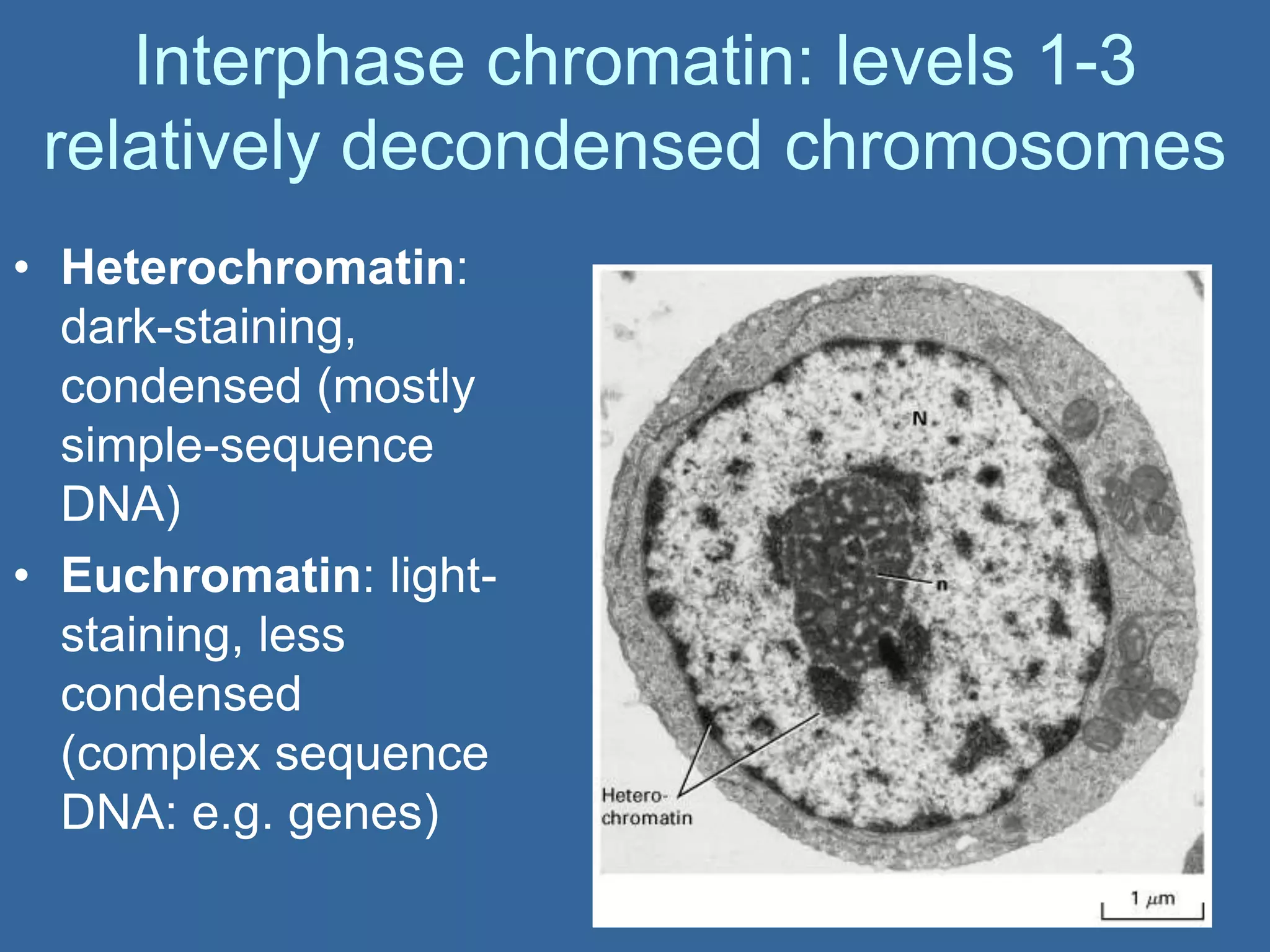
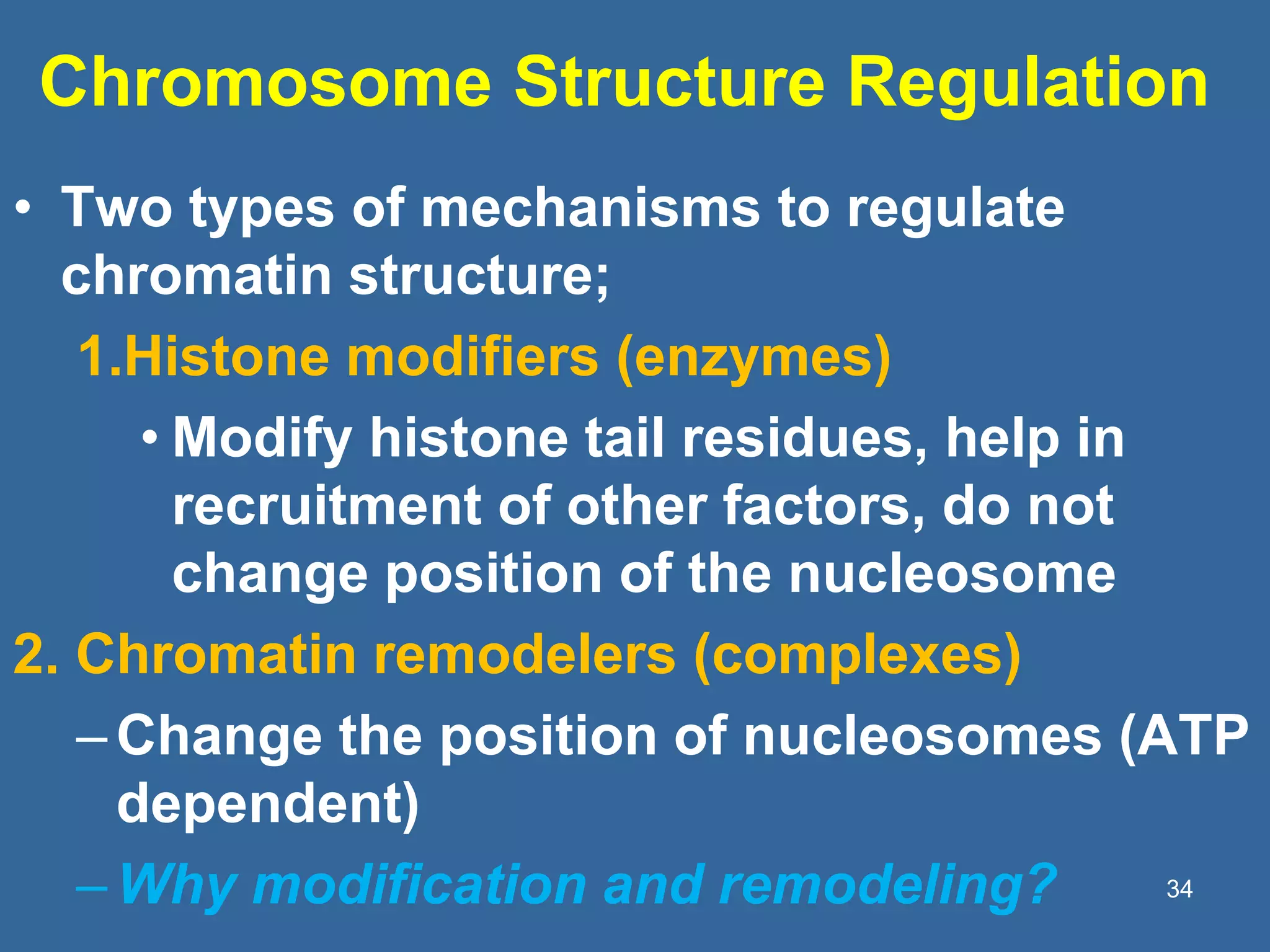
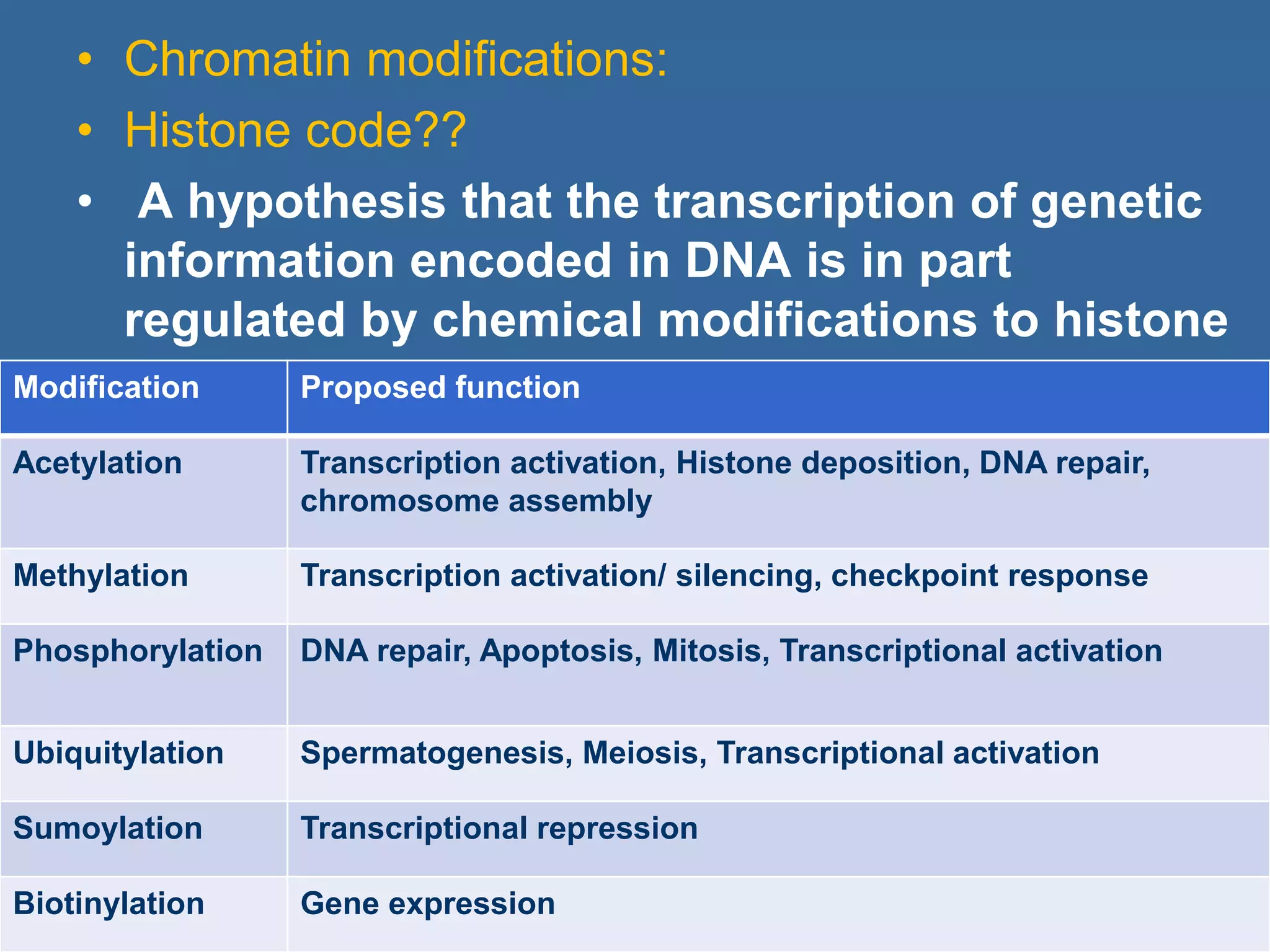
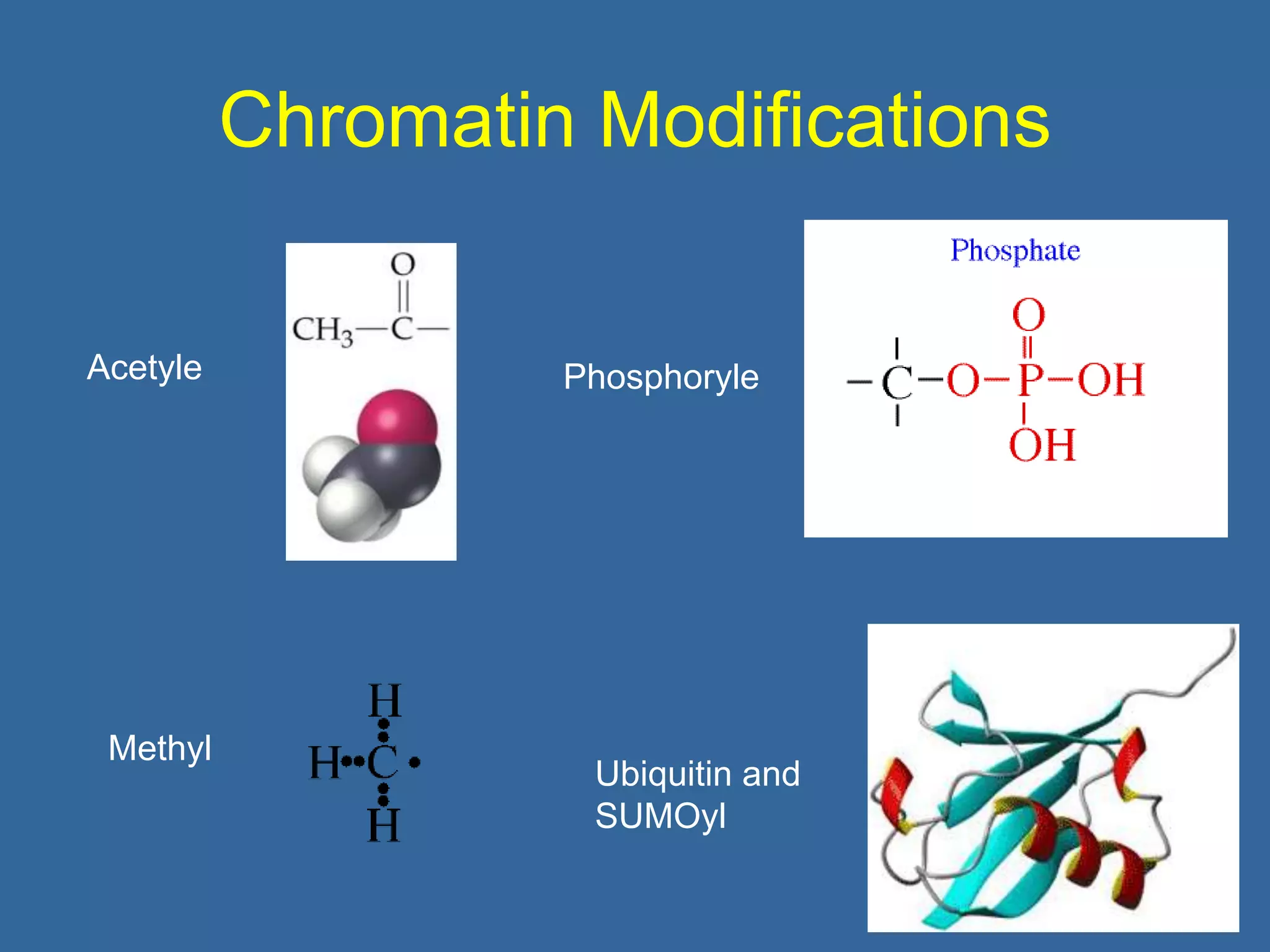
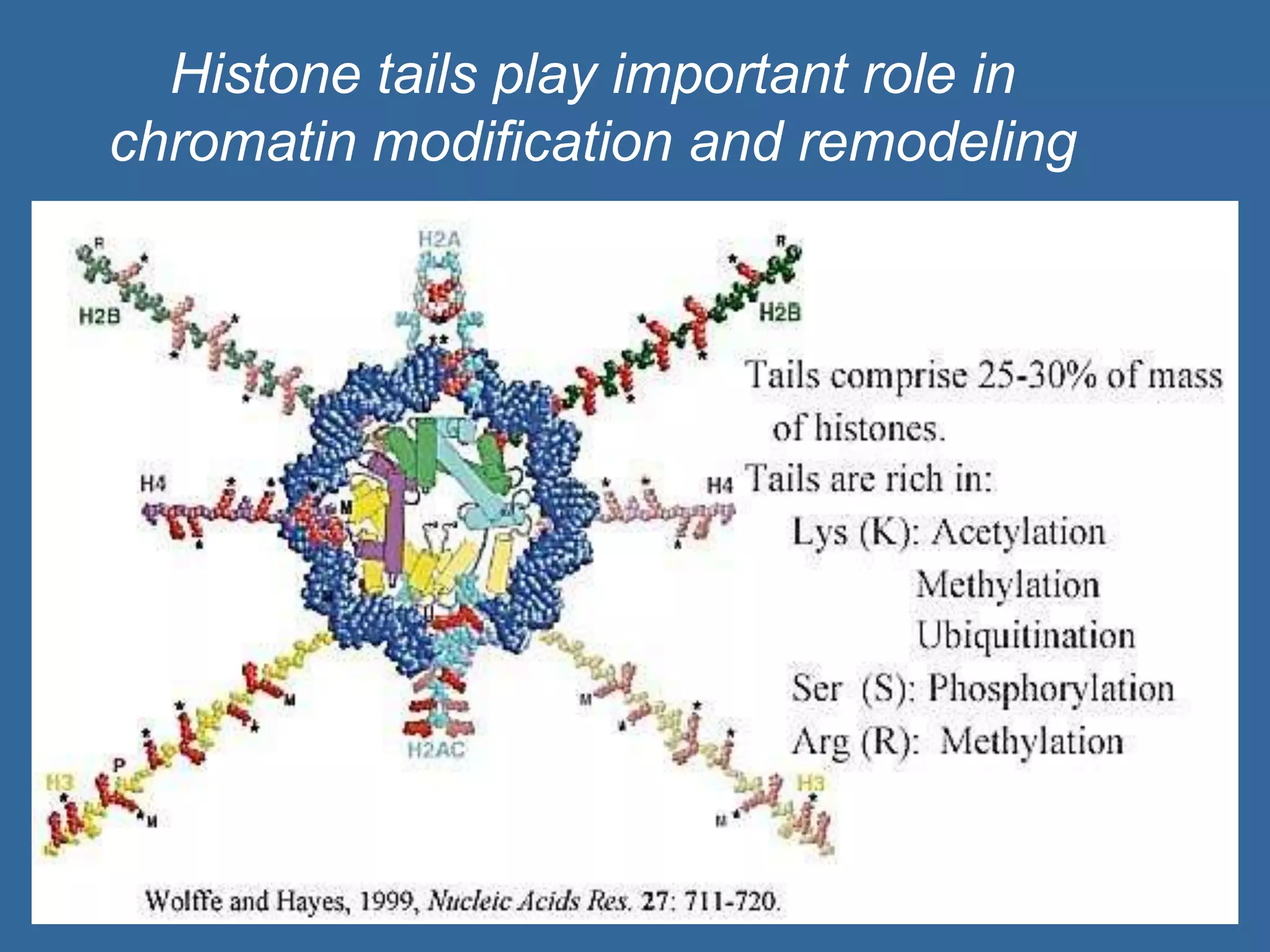
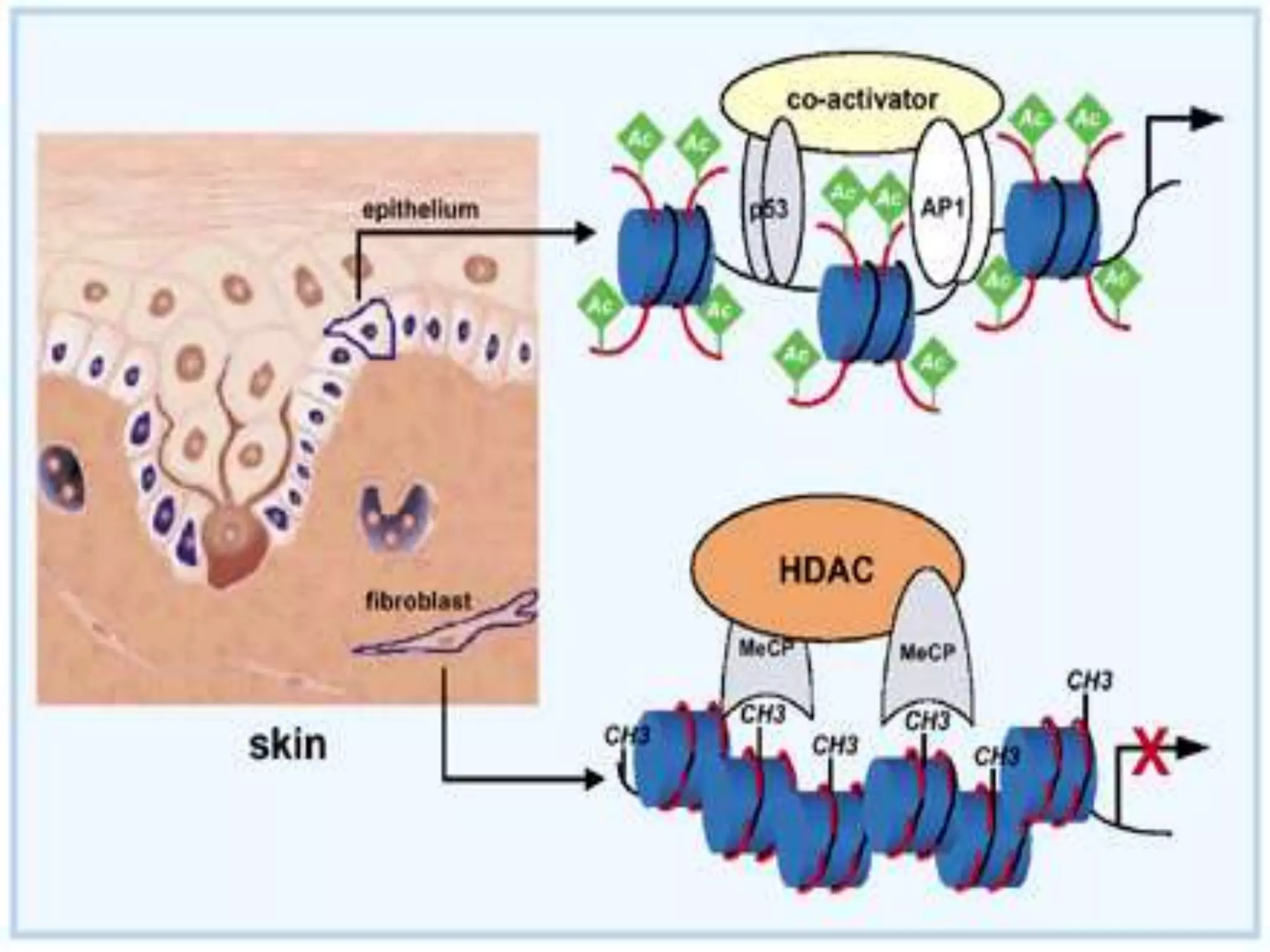
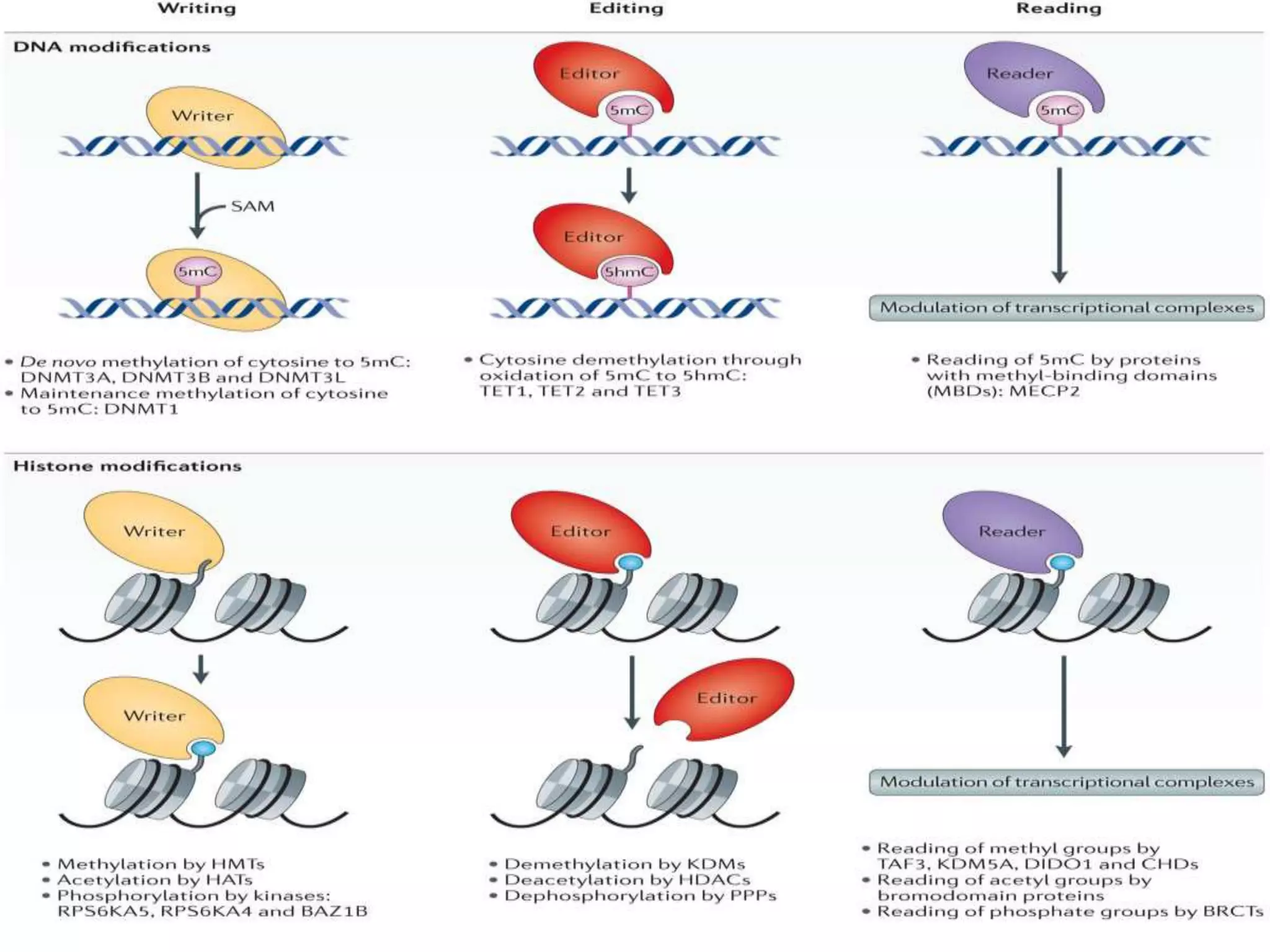
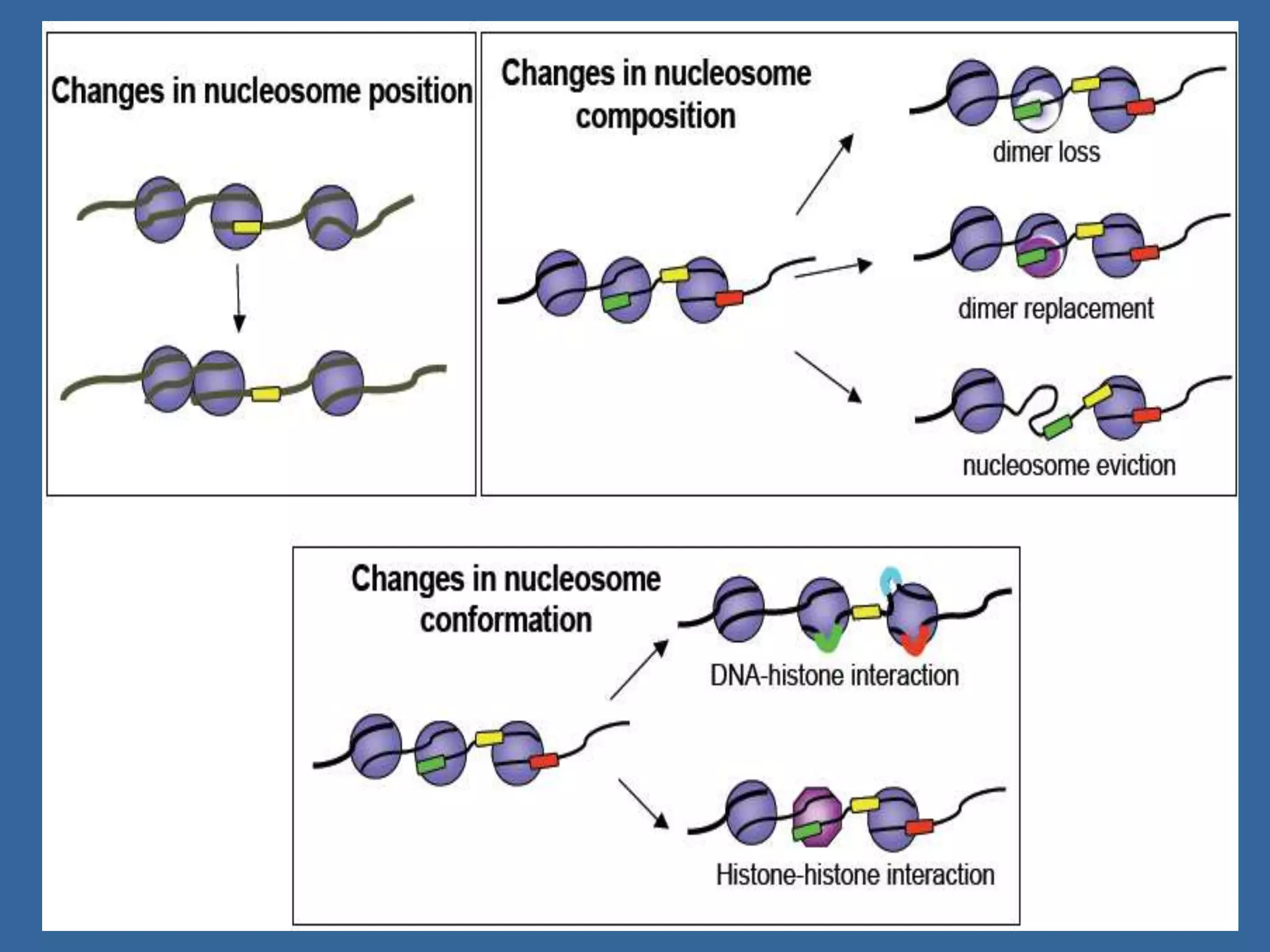
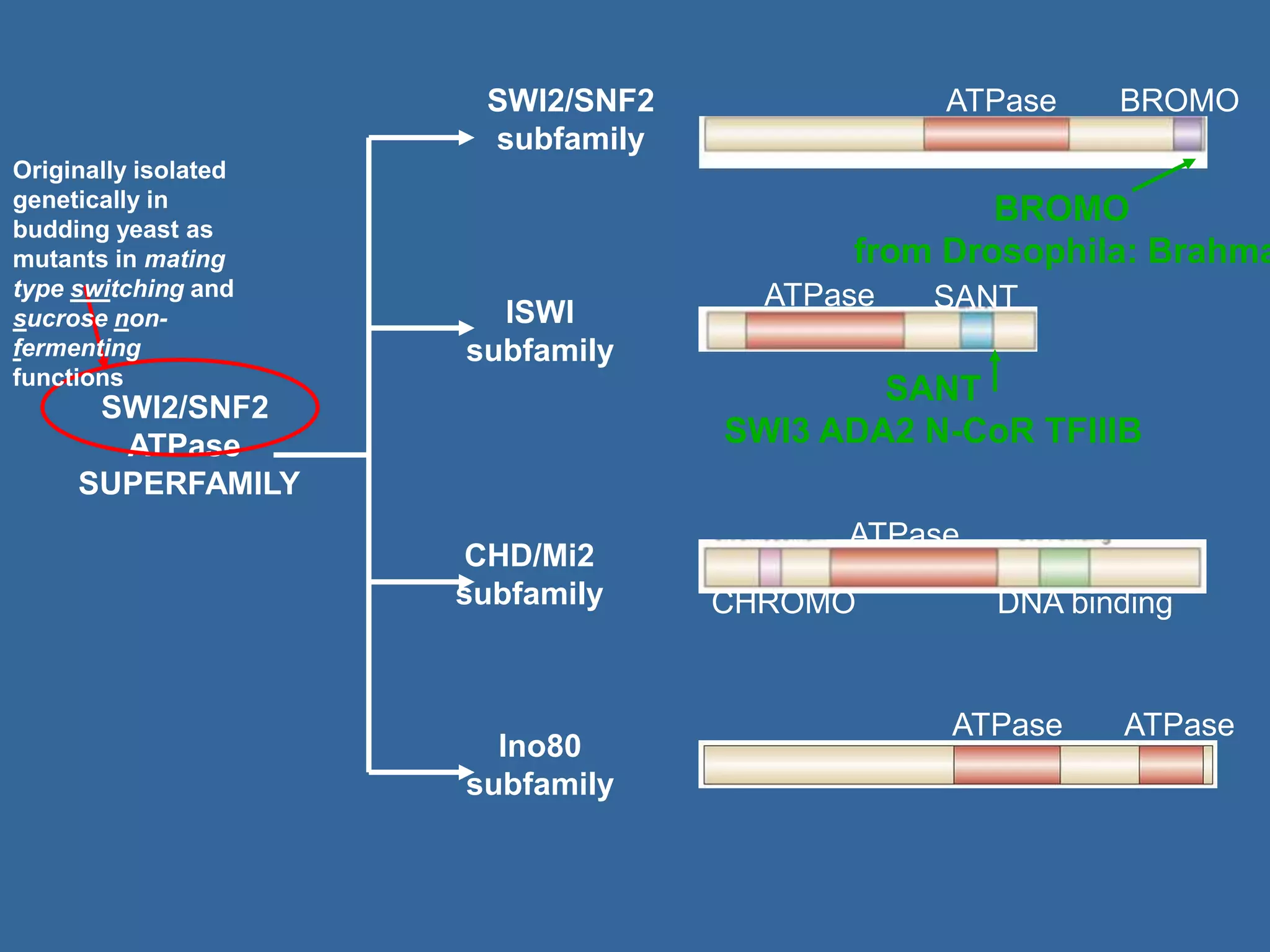
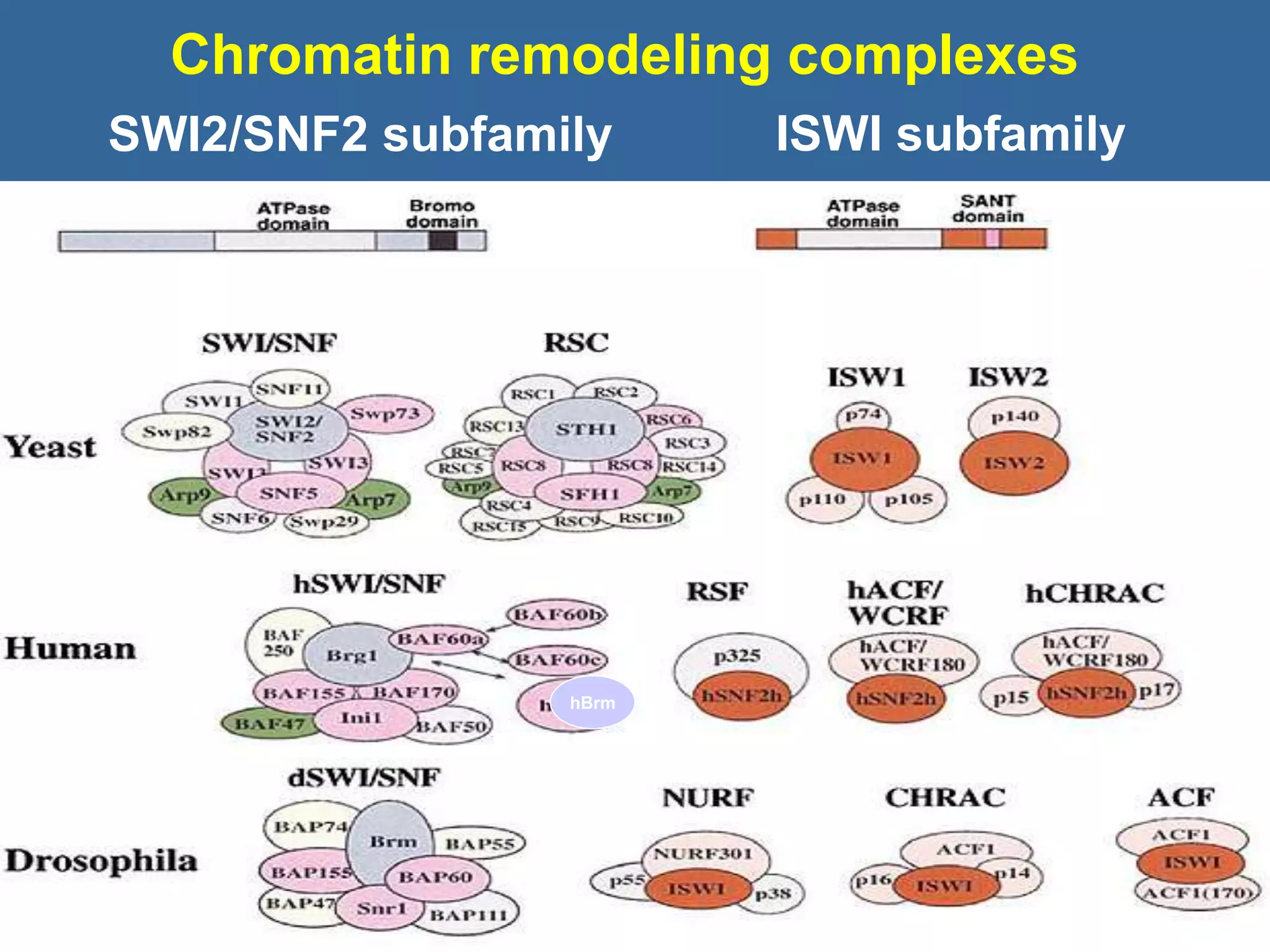
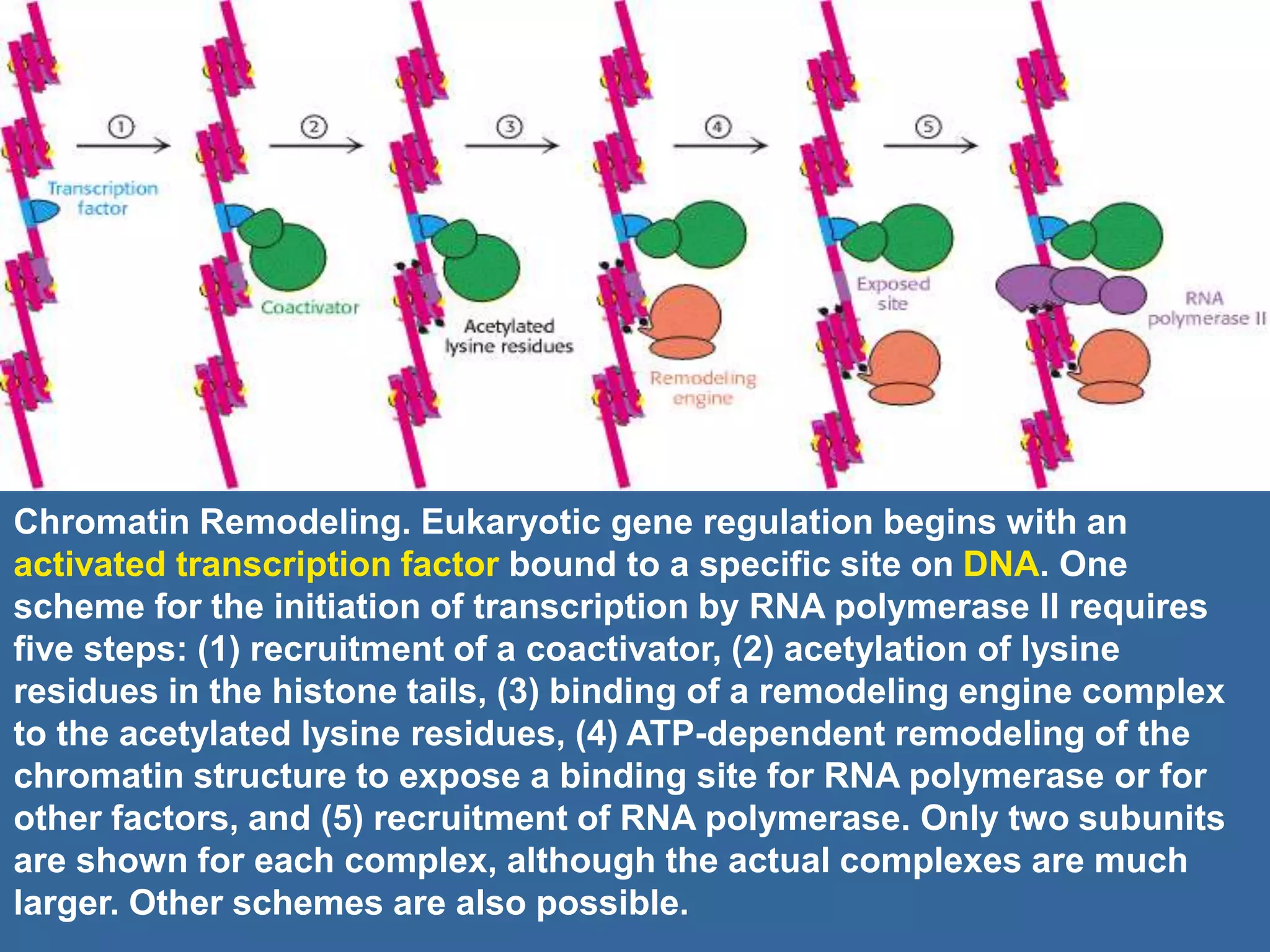
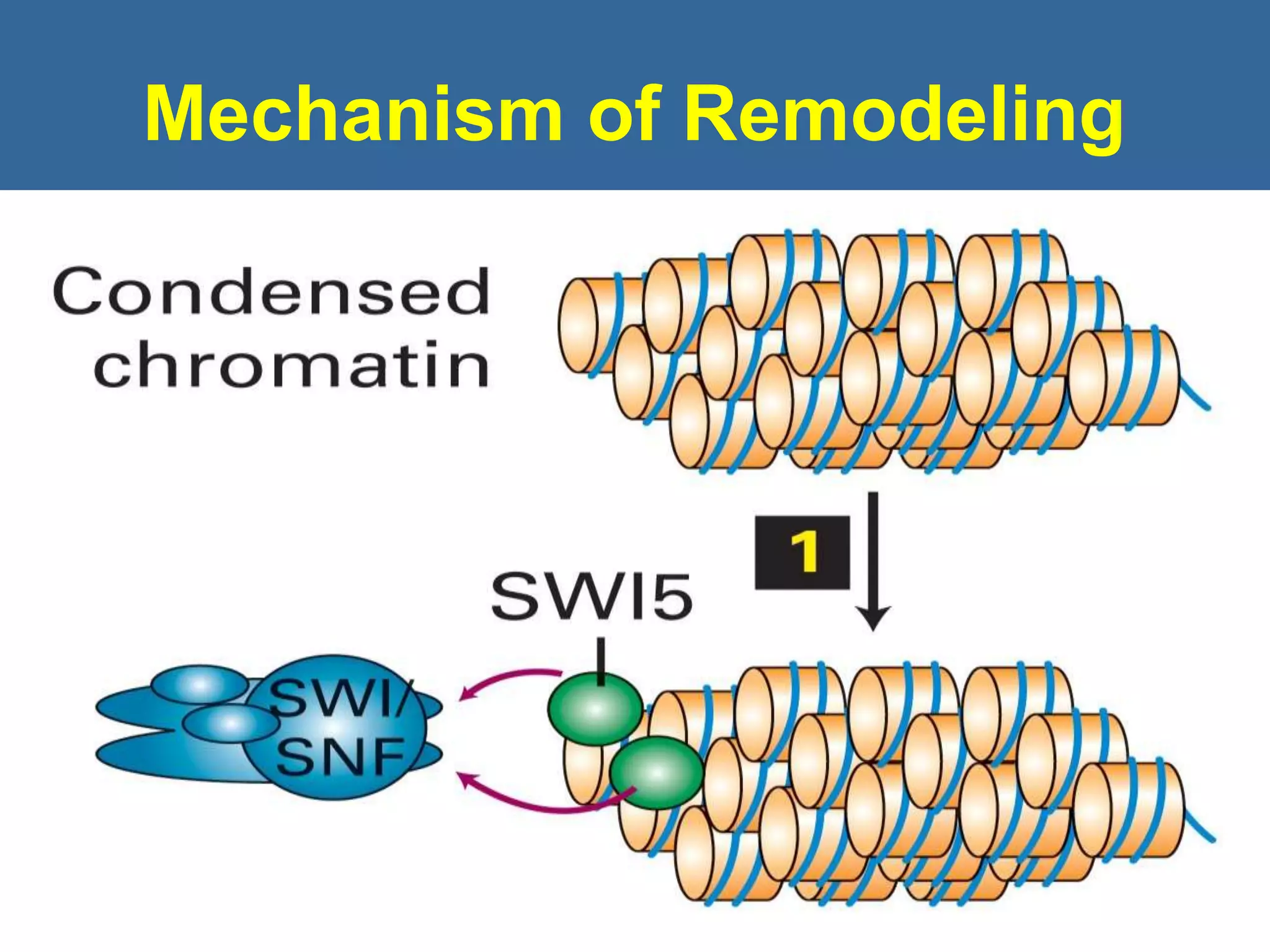
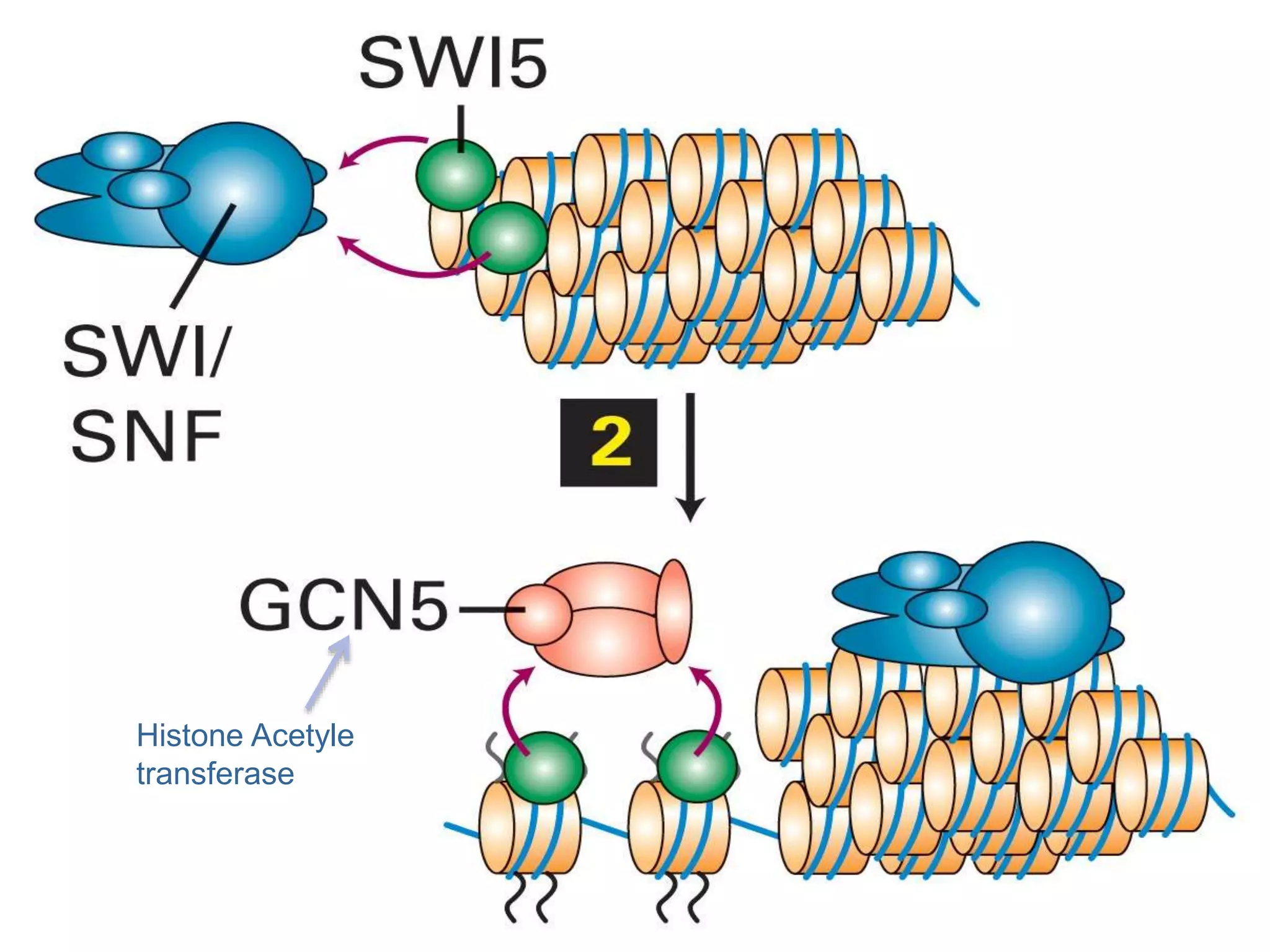
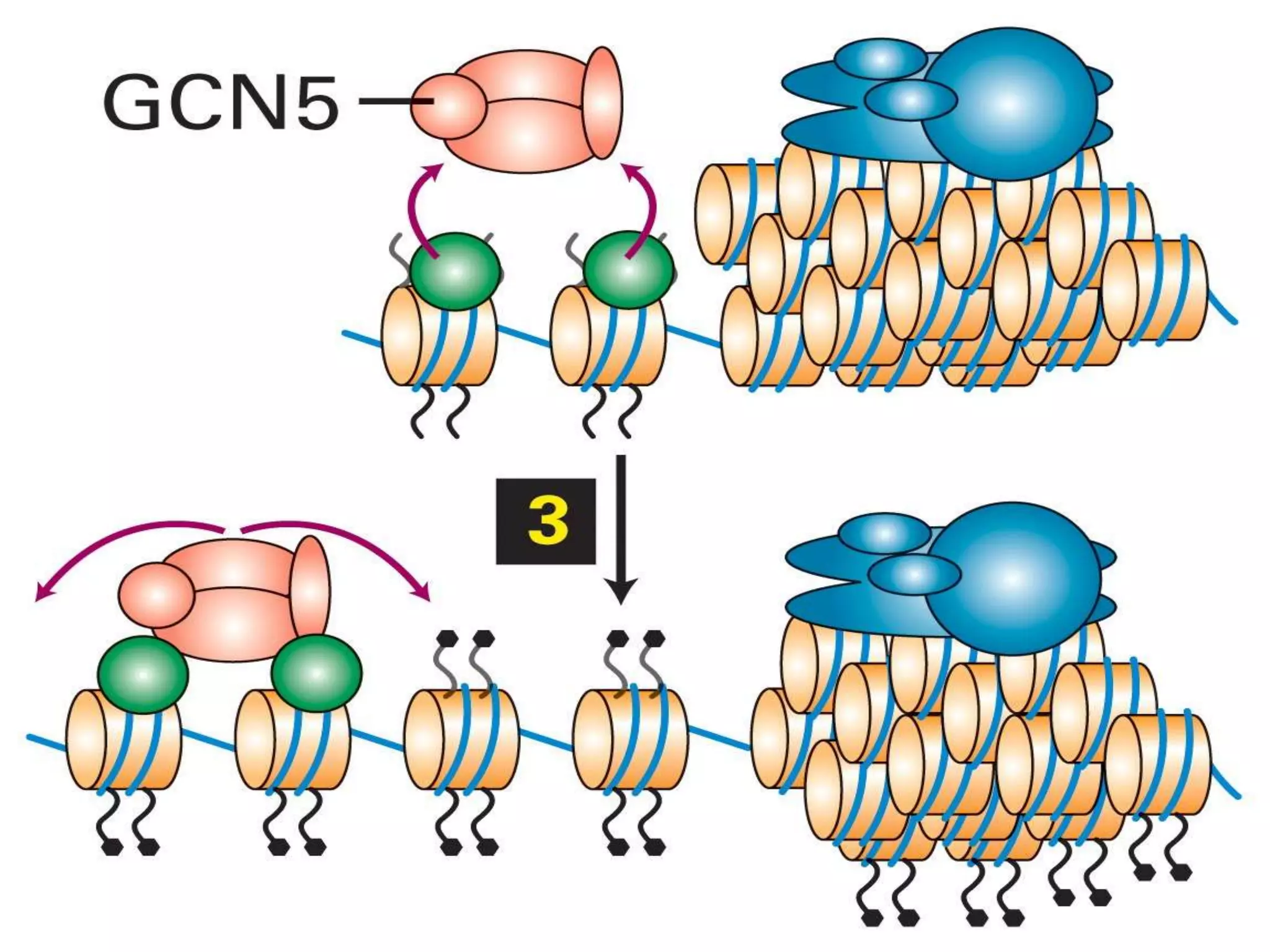
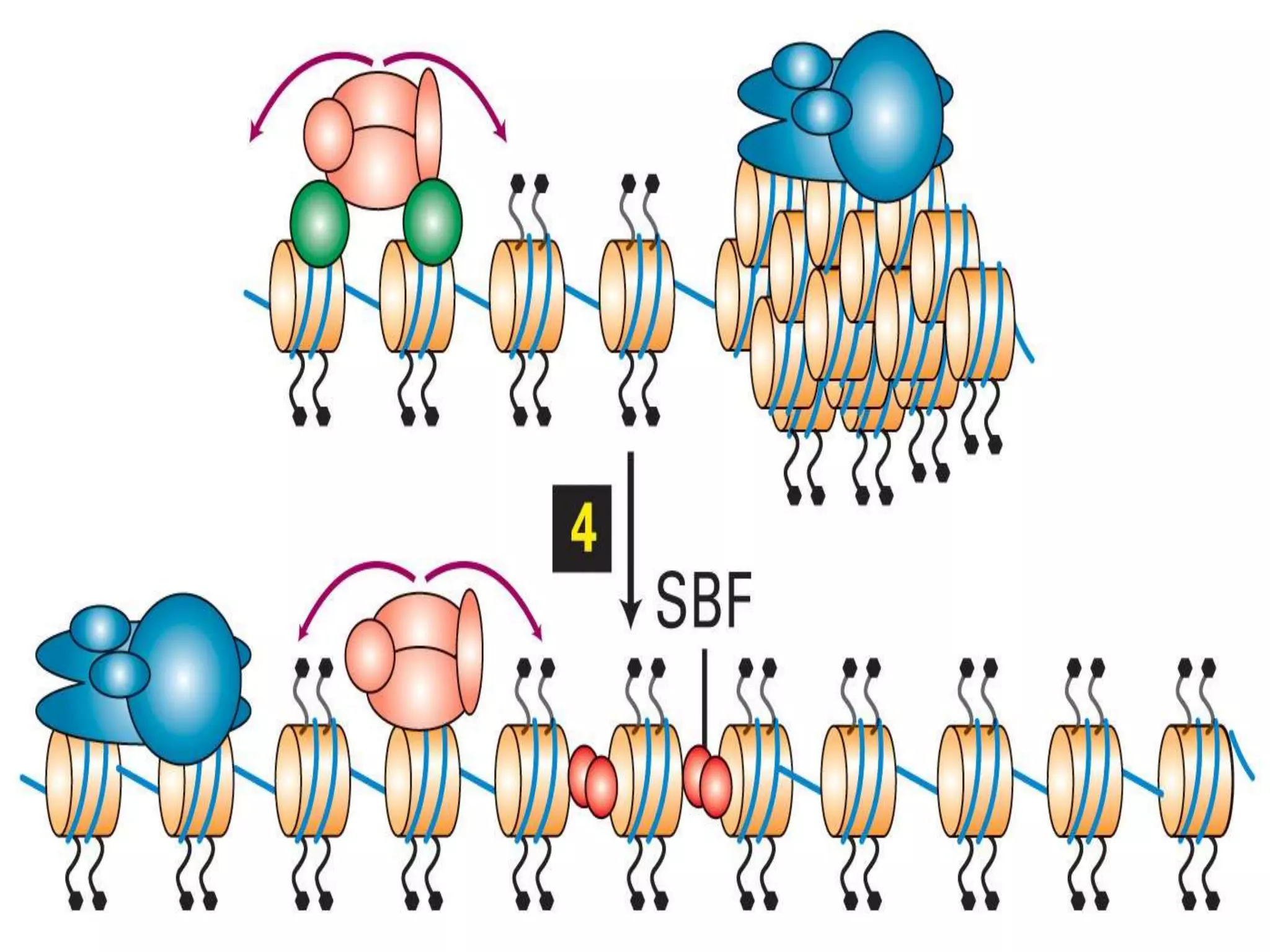
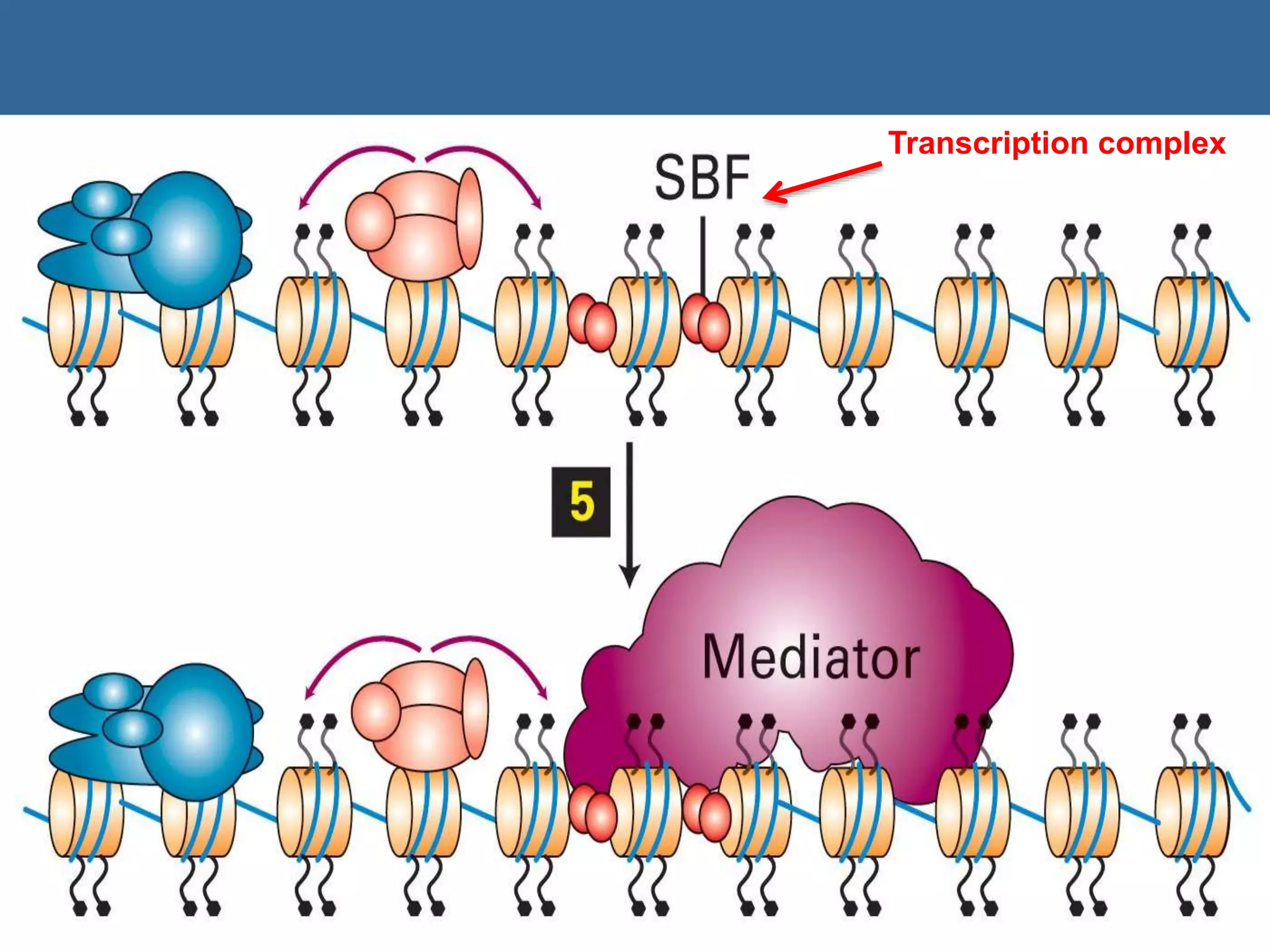
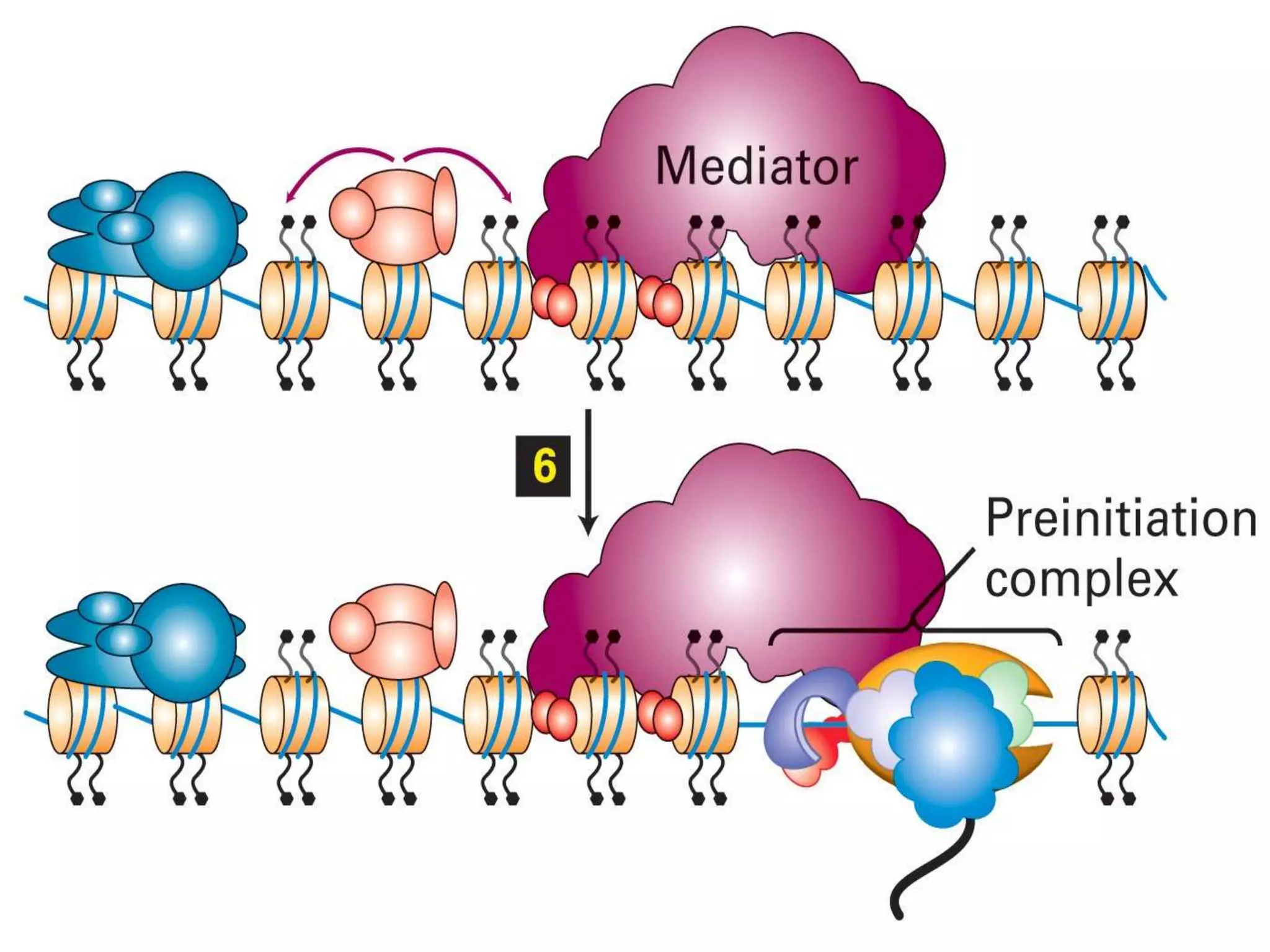
The document details the structural organization of genomes in prokaryotic and eukaryotic cells, emphasizing differences in DNA packaging and complexity. Prokaryotic DNA is compacted via supercoiling and specific proteins, while eukaryotic DNA is organized into nucleosomes around histones, involving multiple levels of chromatin structure. Chromatin modifications and remodeling play critical roles in gene regulation, facilitating access to genetic information and impacting overall cellular functions.