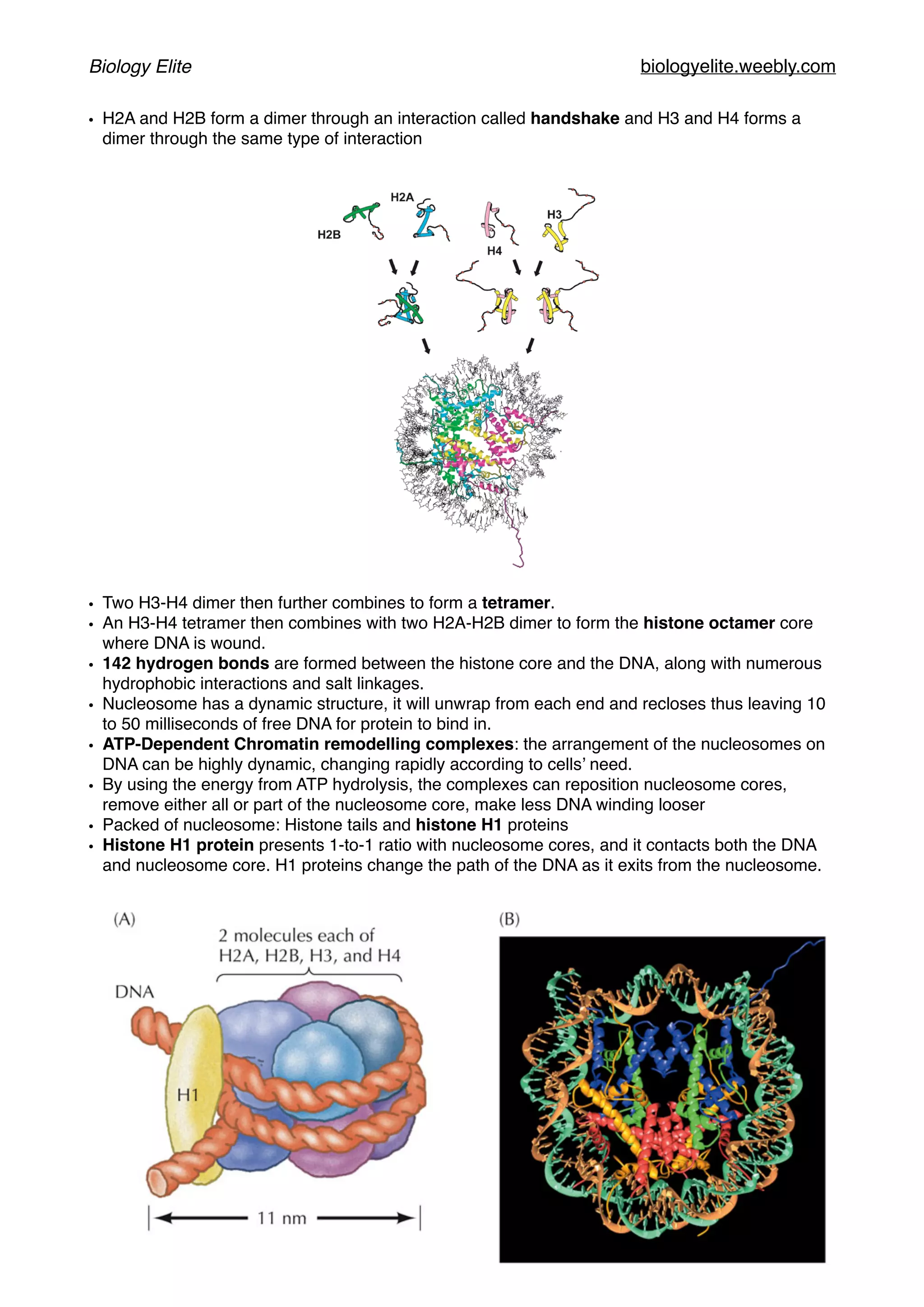
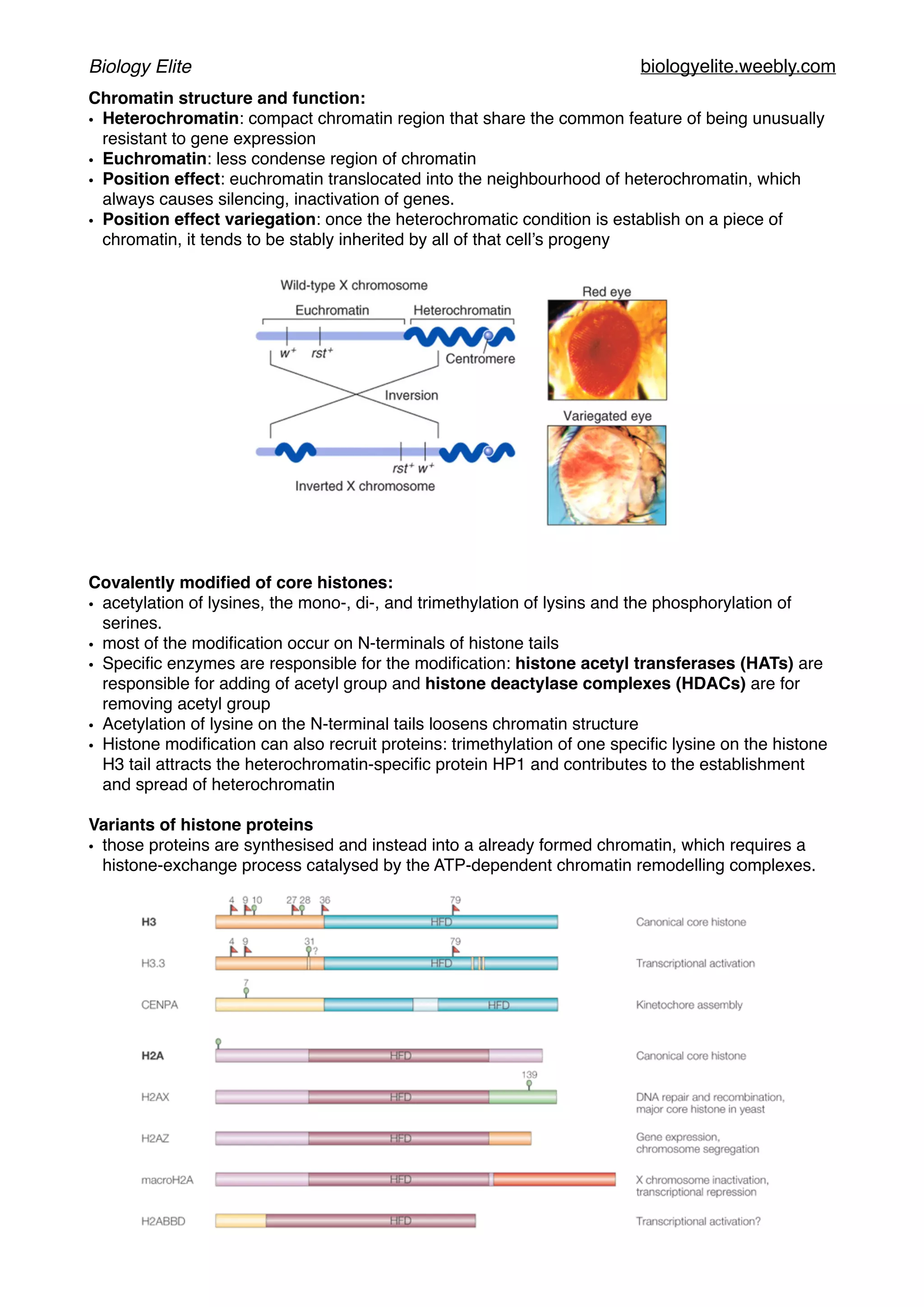
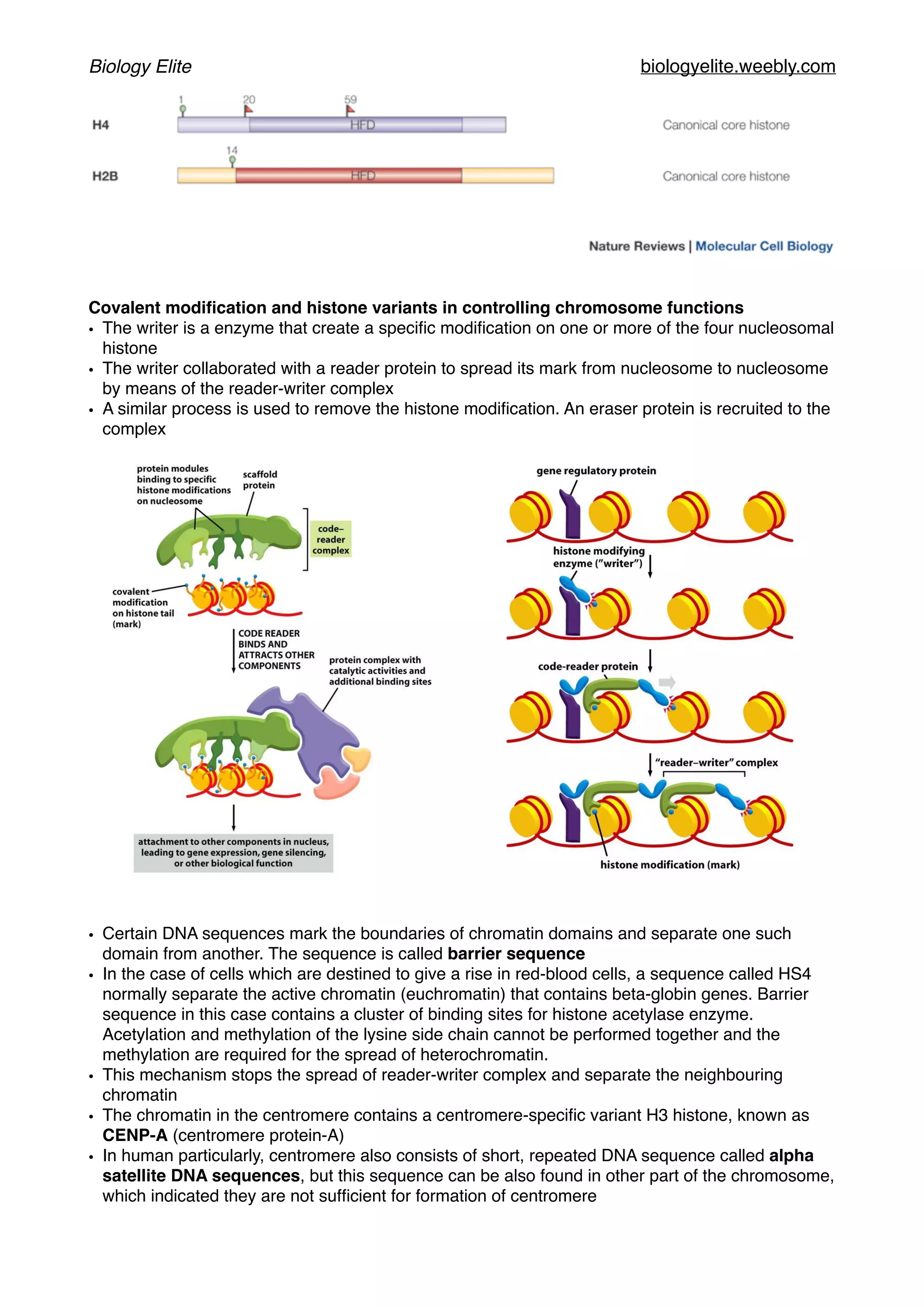
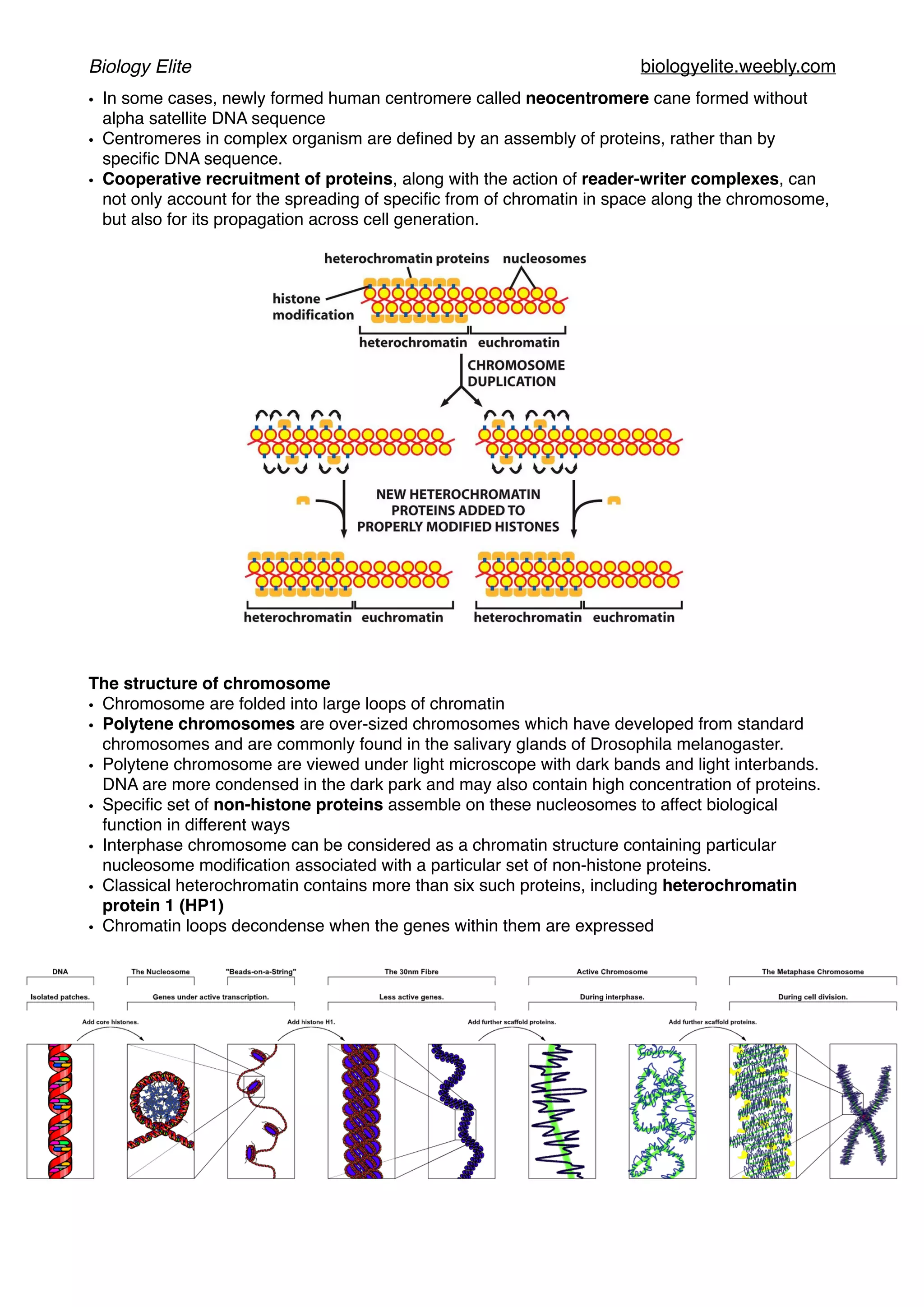
The document discusses the structure and function of DNA, chromosomes, and genomes, explaining the types of base pairings and the organization of DNA into chromosomes, which consist of DNA molecules and proteins. It describes the dynamics of chromatin structure, including the roles of histones, nucleosomes, and chromatin remodeling complexes in gene regulation. Additionally, the document covers genetic evolution, emphasizing the significance of homologous genes, conserved regions, and the mechanisms of genetic variation and selection.