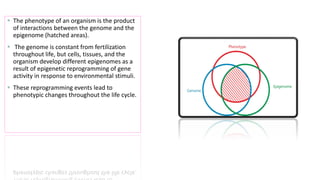
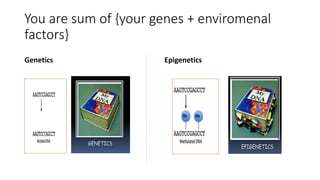
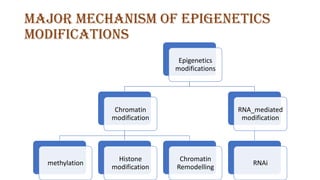
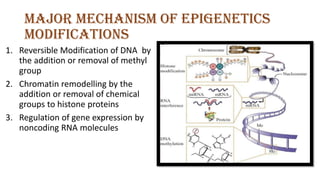
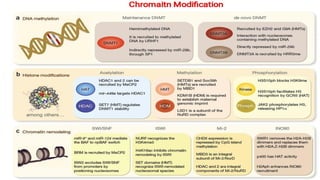
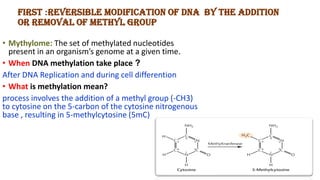
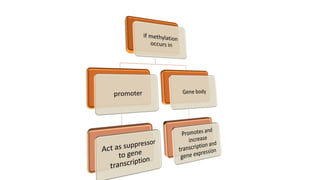
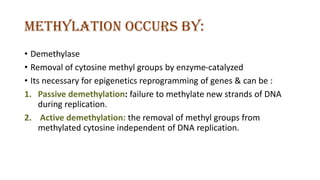
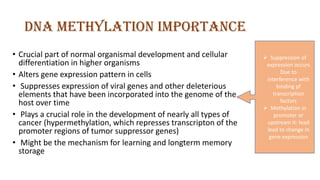
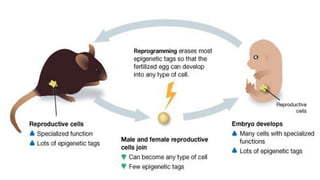
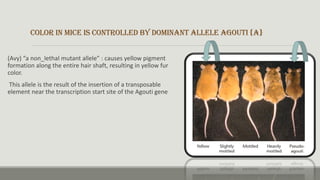
The document discusses epigenetics, focusing on mechanisms that regulate gene expression without altering the DNA sequence itself, including DNA methylation and histone modification. It highlights how epigenetic changes can be heritable and affect phenotypic traits across generations, exemplified by environmental influences like diet and trauma. Specific case studies, such as the impact of childhood trauma on gene demethylation and color variations in mice, illustrate the significance of epigenetic inheritance in various biological contexts.