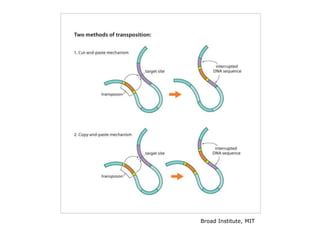
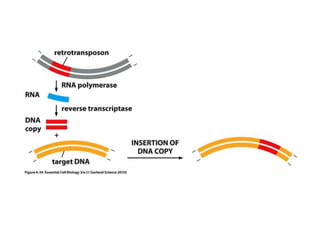
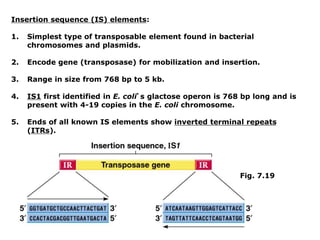
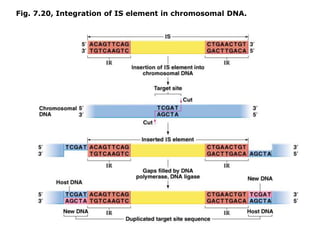
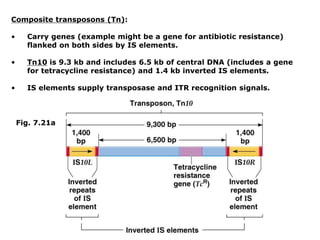
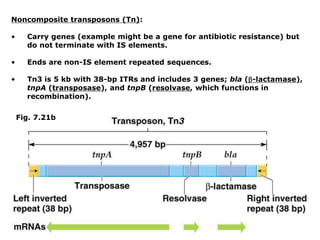
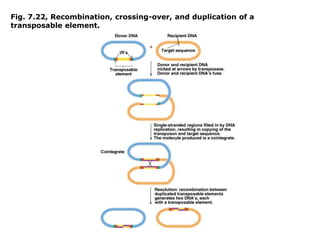
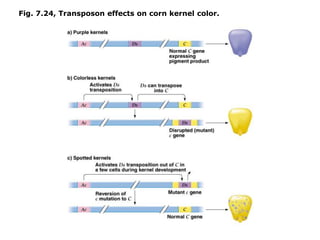
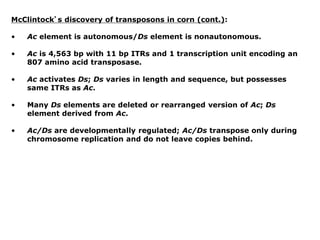
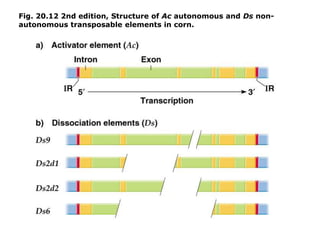
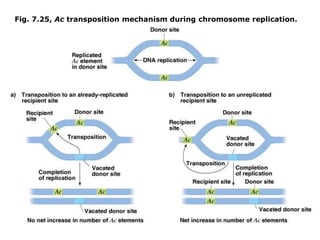
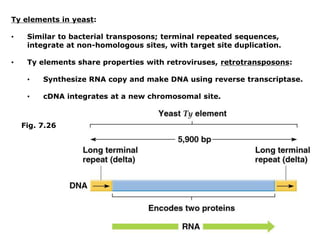
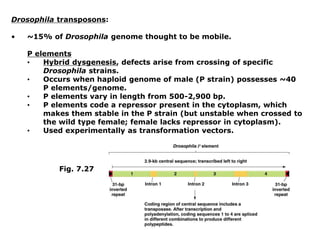
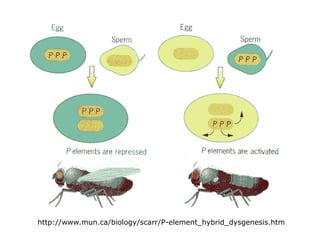
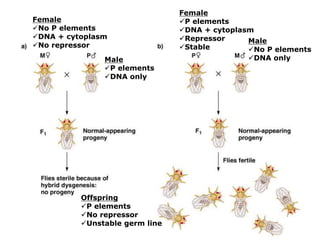
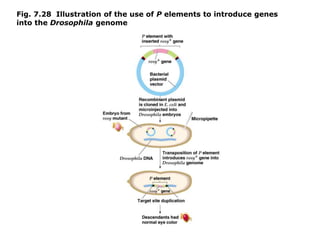
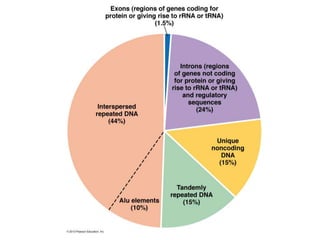
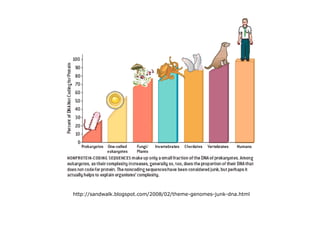
Chapter 7b discusses transposable elements, which are mobile genetic components in prokaryotic and eukaryotic genomes that can relocate within the genome, leading to genetic mutations and contributing to evolution. It covers two types of transposable elements: insertion sequences and transposons, detailing their mechanisms of movement such as cut-and-paste and copy-and-paste, as well as their effects on gene expression and genomic structure. The chapter also highlights significant discoveries by Barbara McClintock regarding plant transposons and their role in kernel color variations in corn.