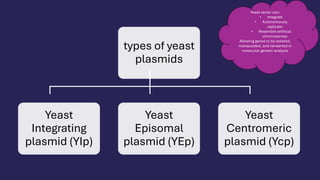
The document discusses the challenges and methods involved in gene transfer and cloning, particularly in eukaryotic systems, highlighting issues with expression in bacteria and the need for alternative expression systems. It details the process of constructing cDNA libraries, oligonucleotide probes, and various types of yeast plasmids used in genetic engineering. Additionally, it addresses properties of shuttle vectors and applications of yeast as a model for recombinant DNA technology.