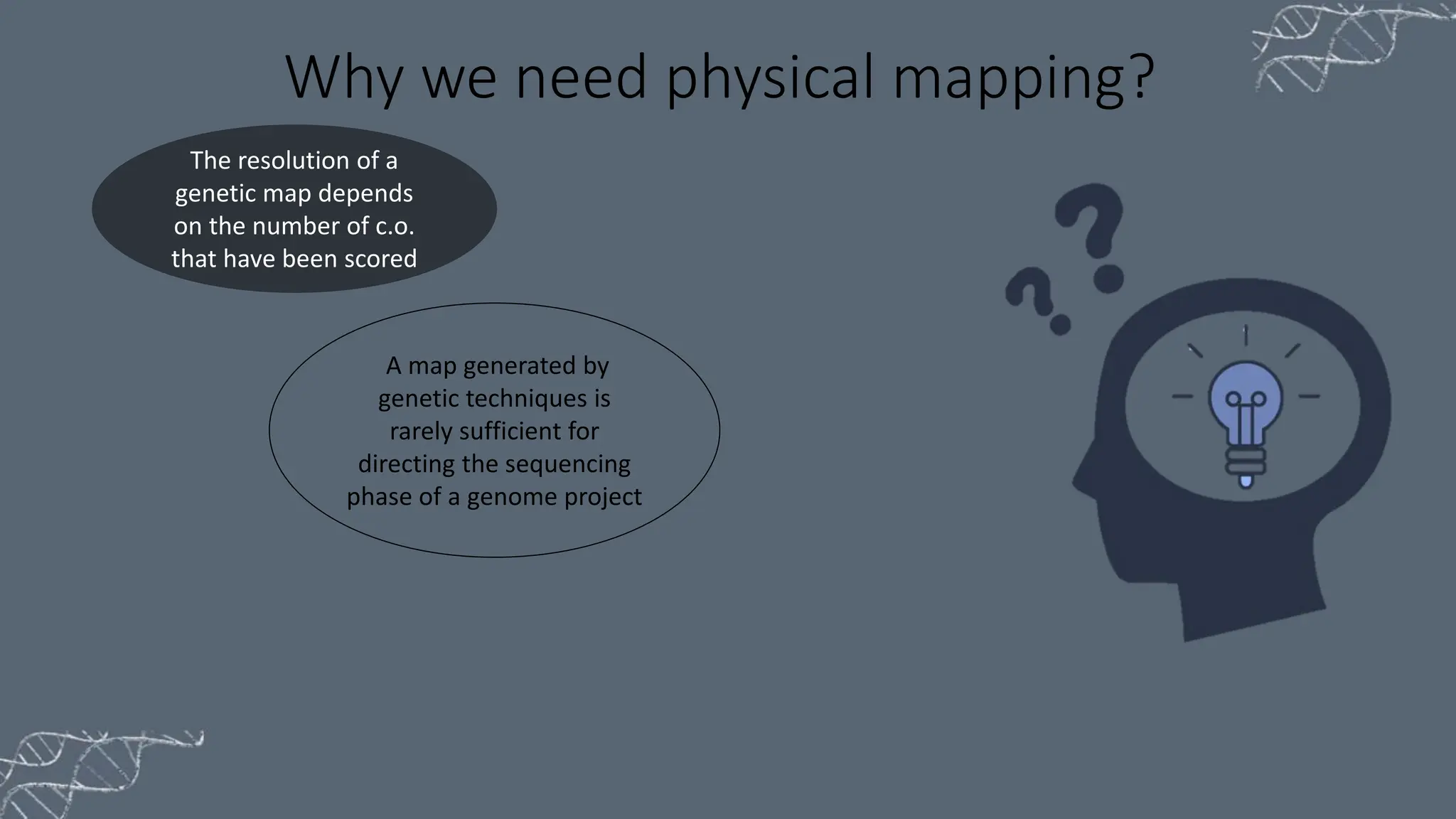
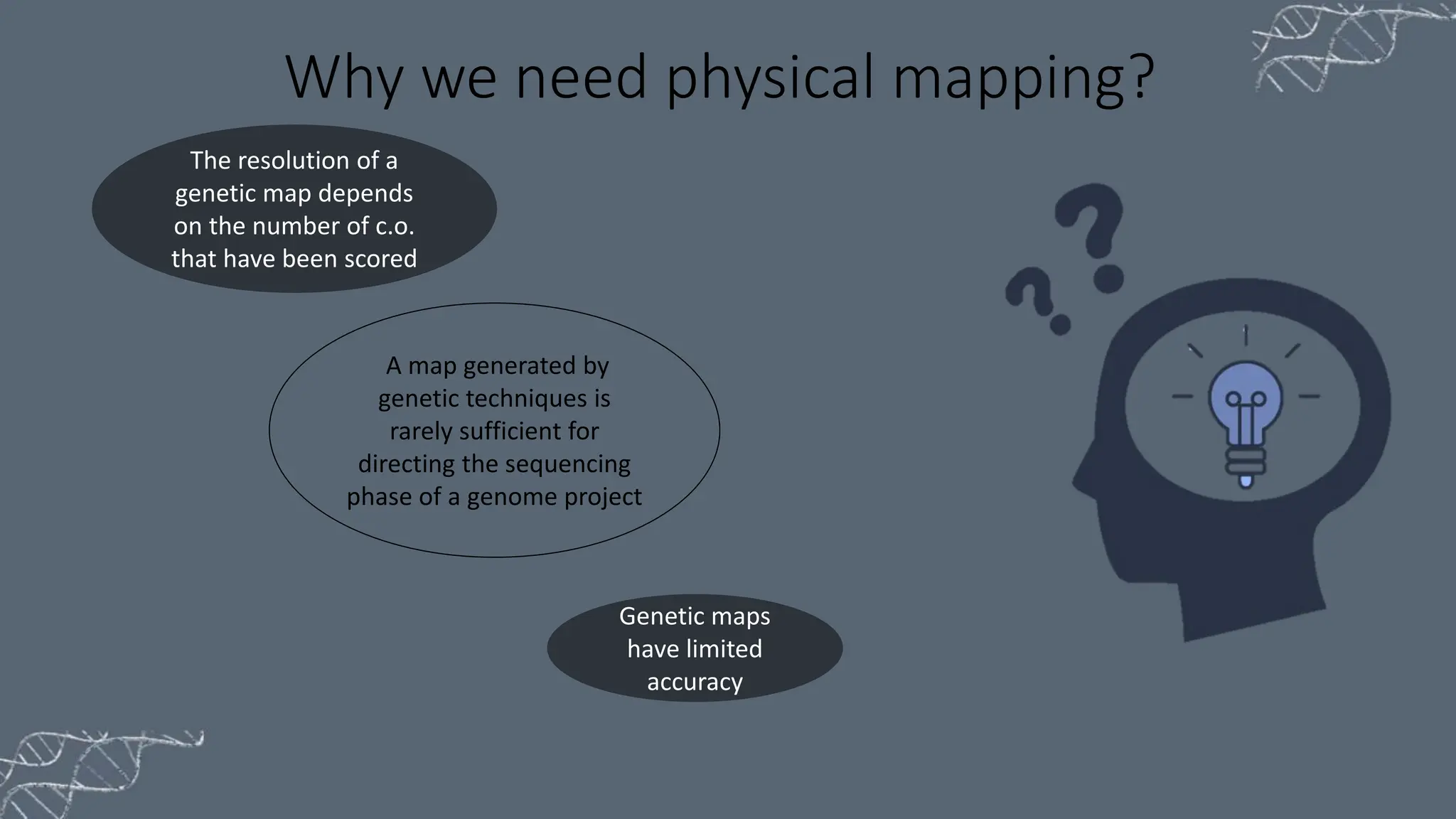
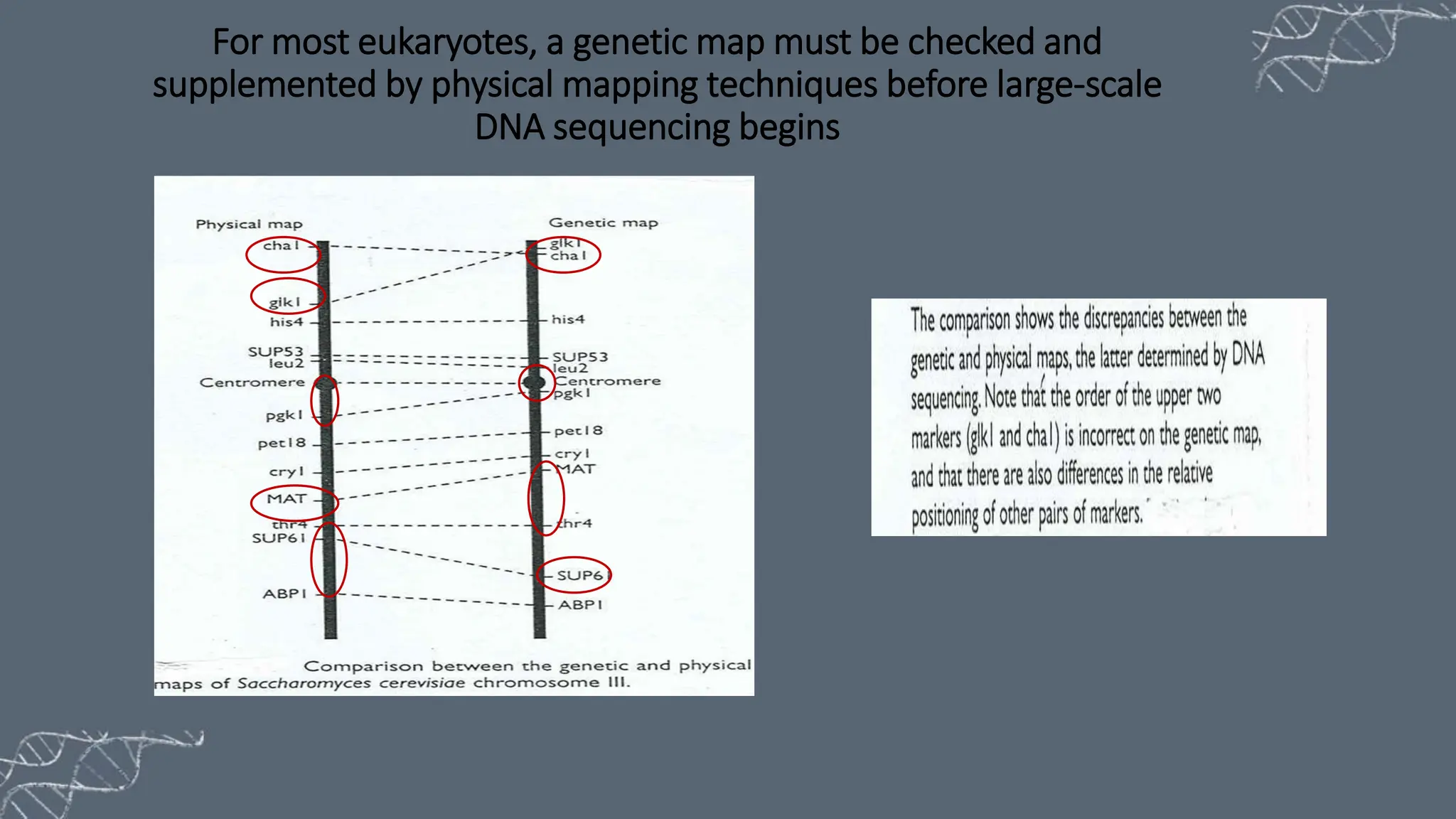
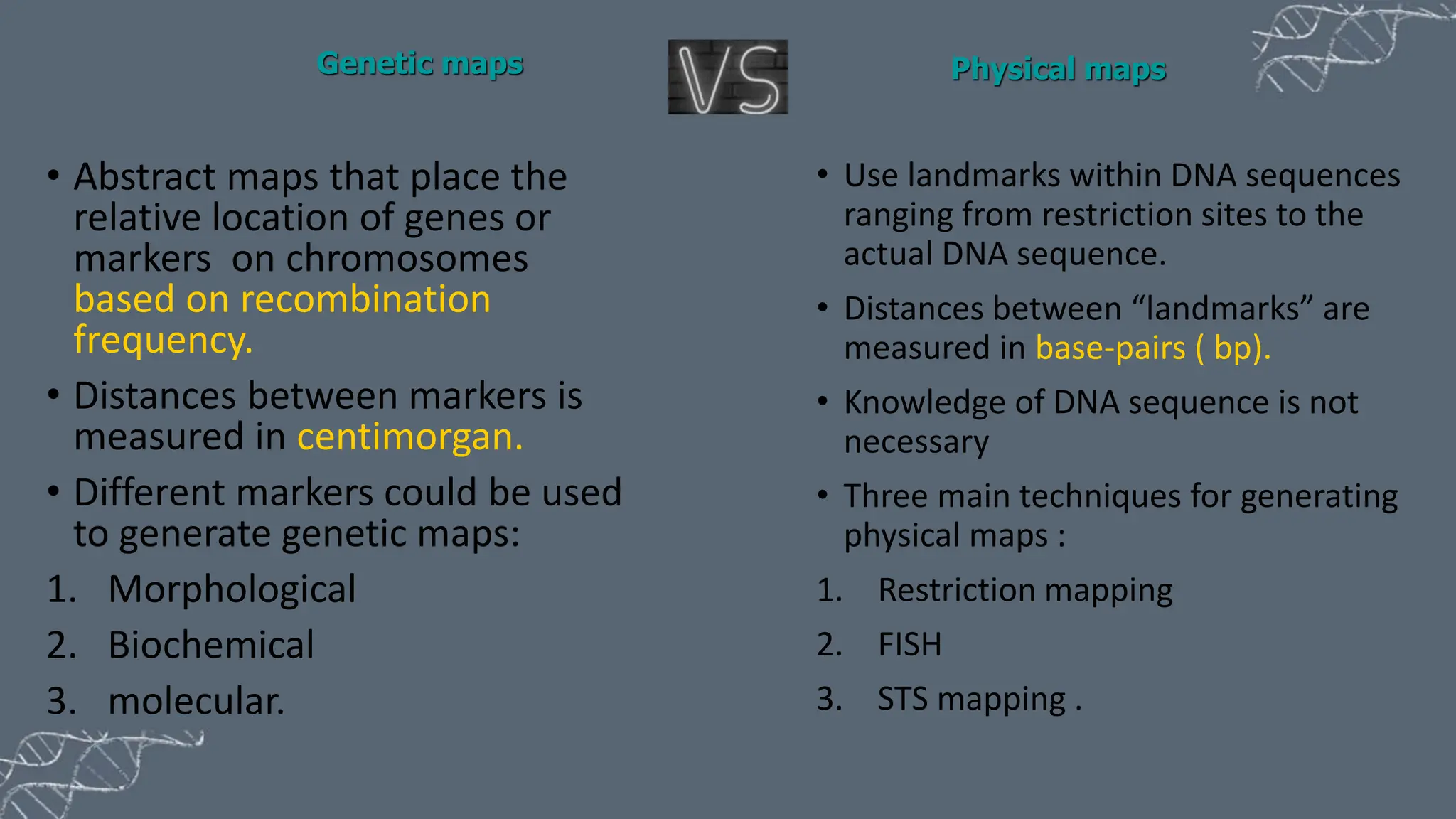
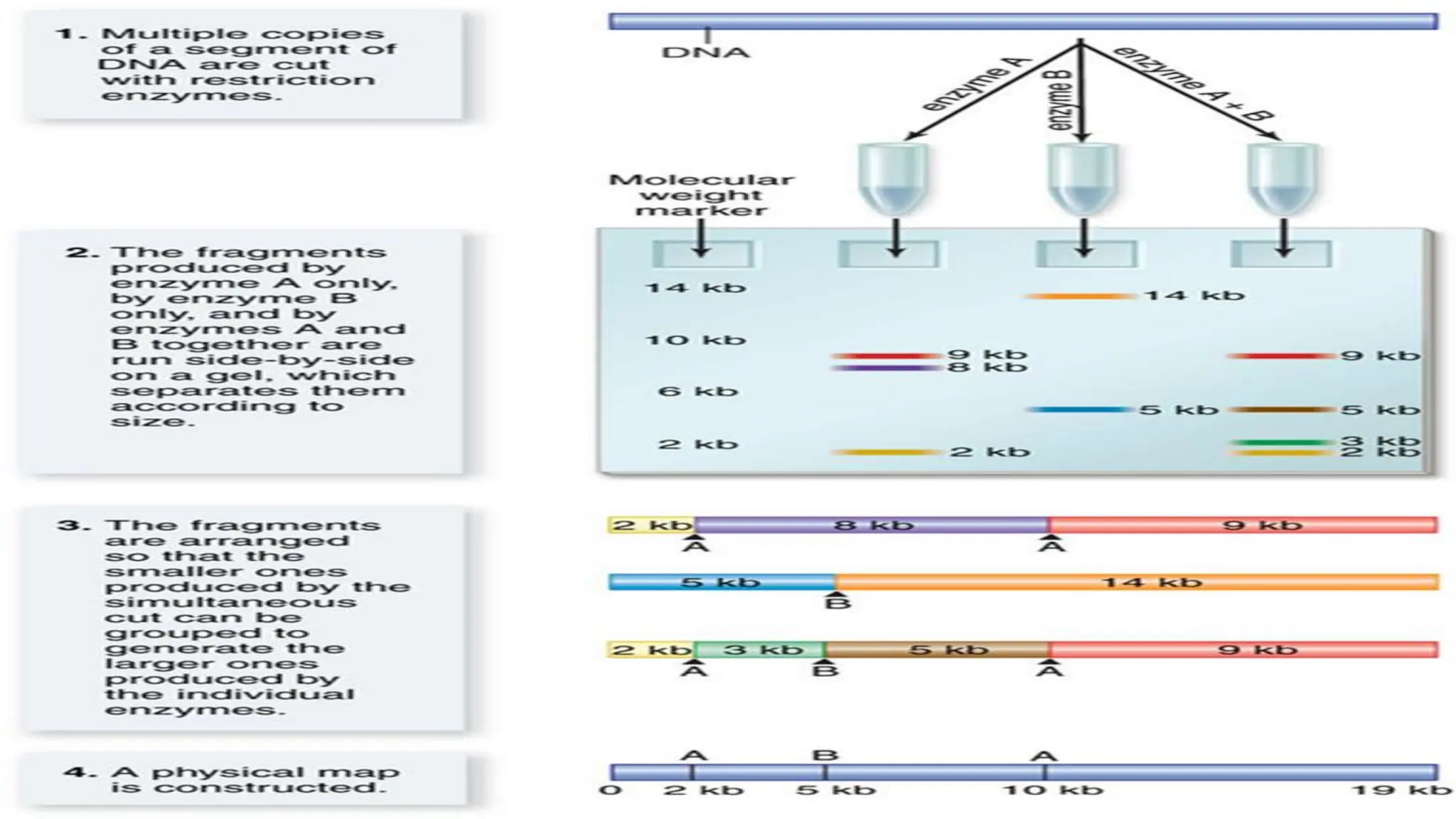
The document discusses the necessity of physical mapping in genome projects, highlighting the limitations of genetic maps that are often insufficient for sequencing direction due to resolution issues and inaccuracies. It describes physical maps, which utilize DNA sequence landmarks and are created using techniques like restriction mapping, FISH, and STS mapping. Additionally, it explains the methods and challenges associated with restriction mapping and pulsed field gel electrophoresis (PFGE) for handling large DNA fragments.