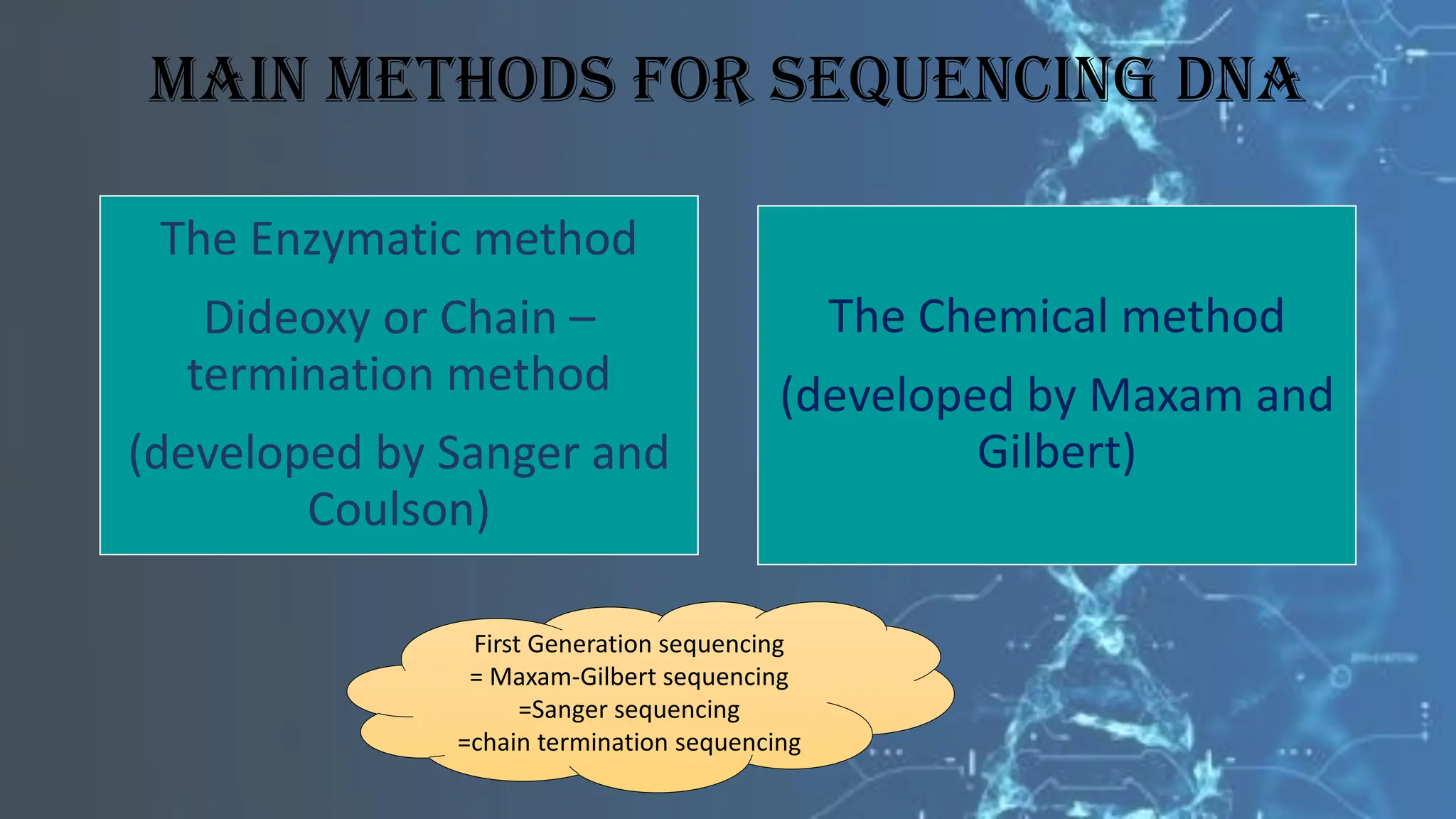
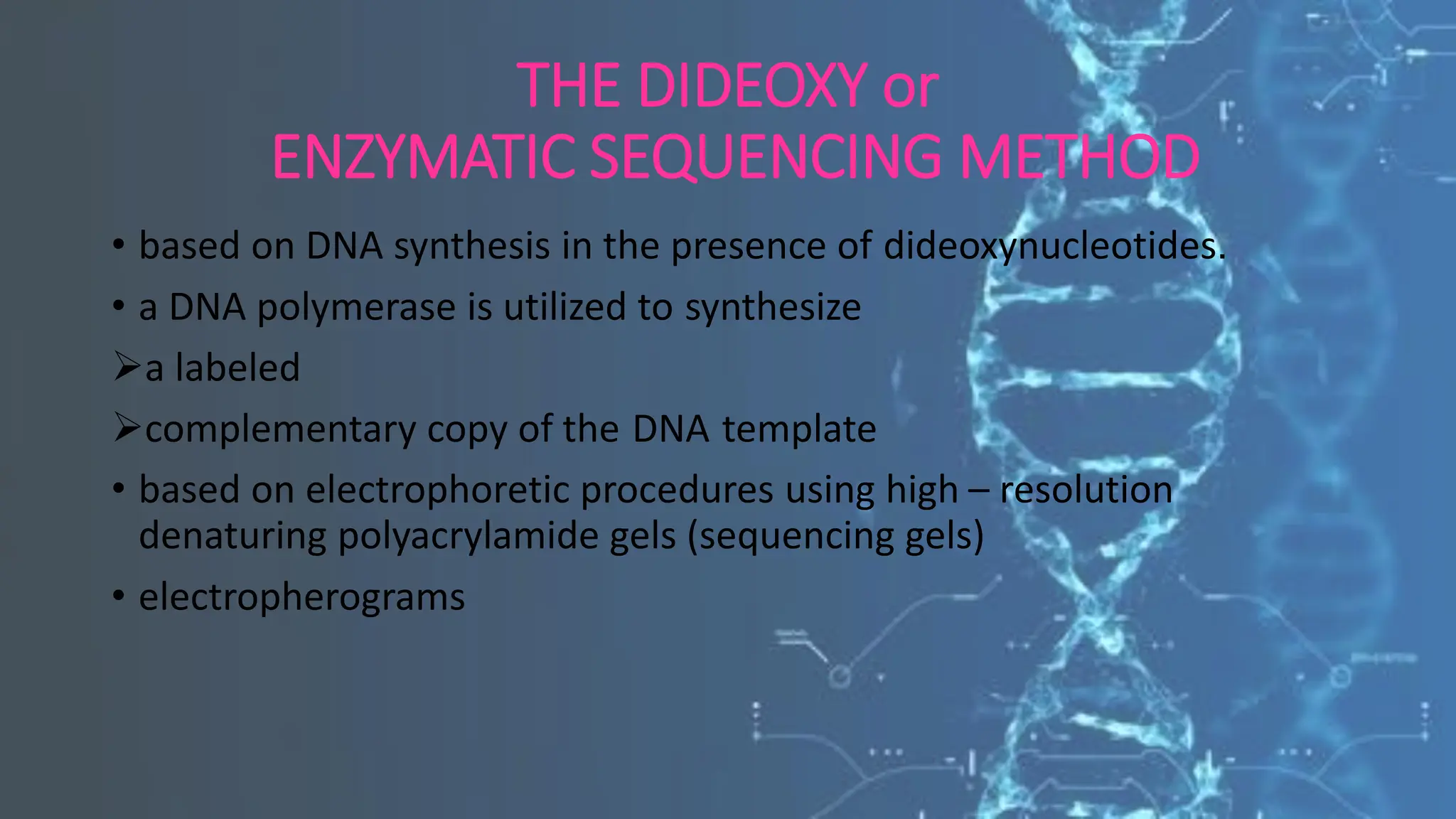
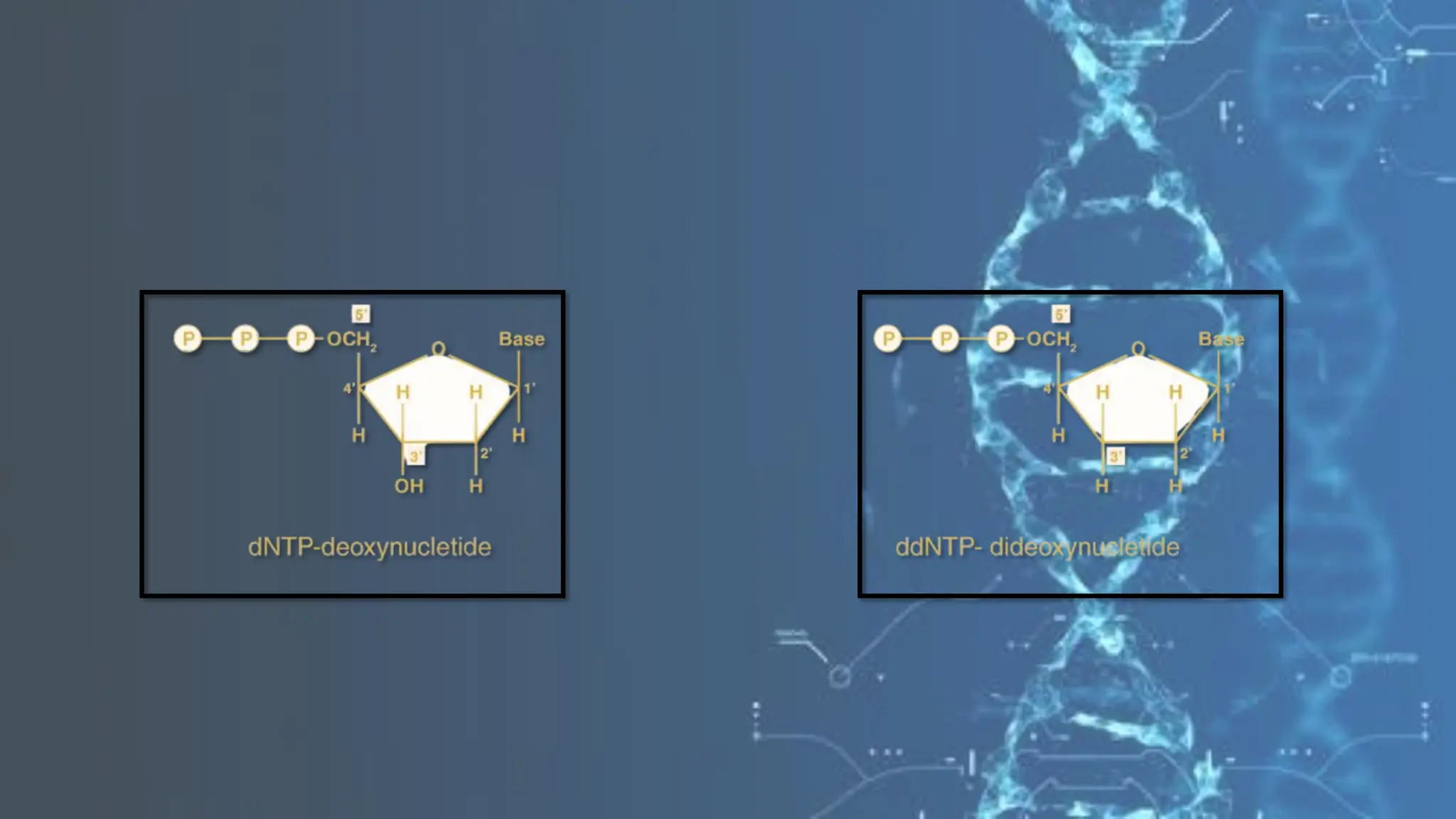
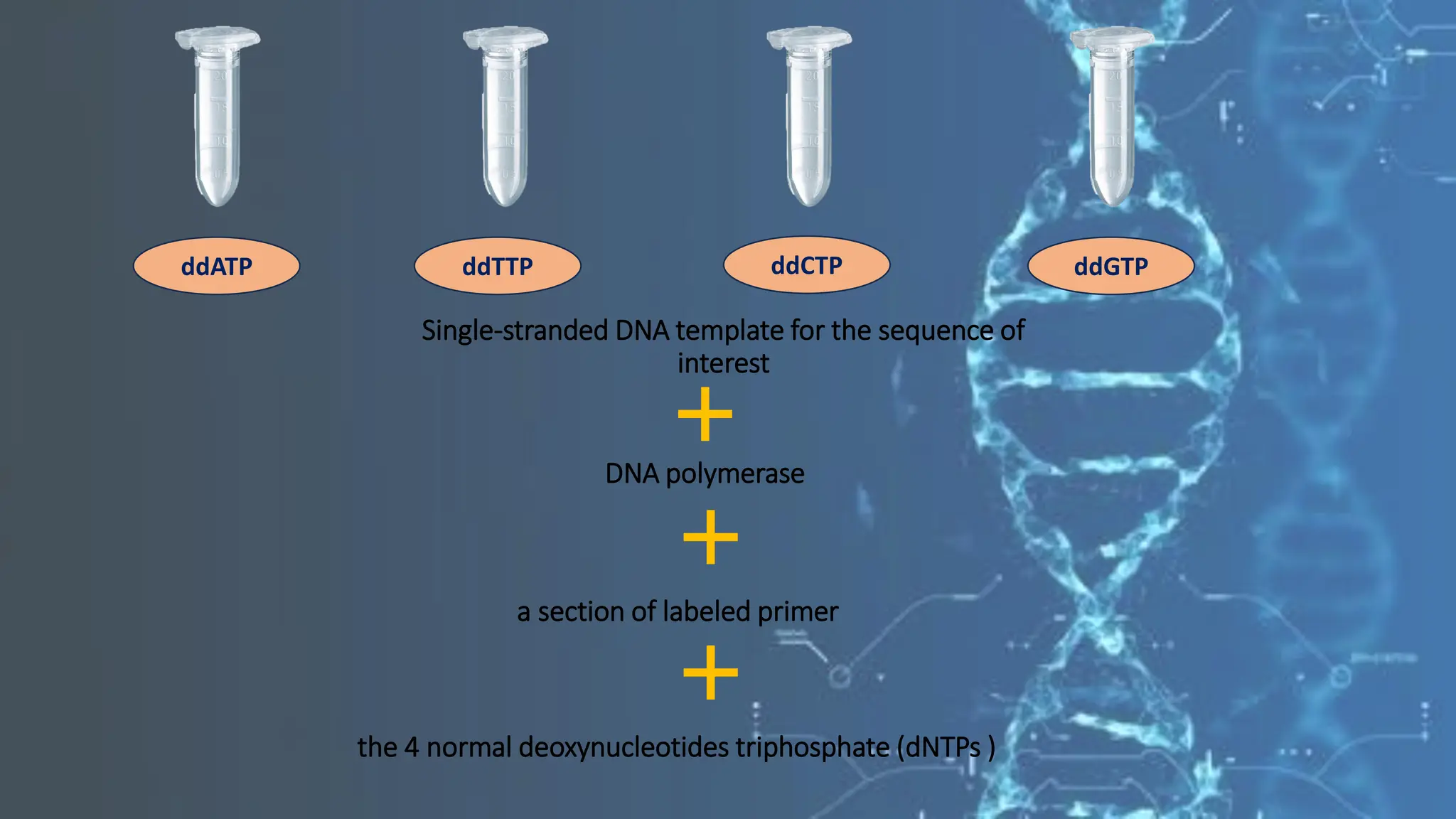
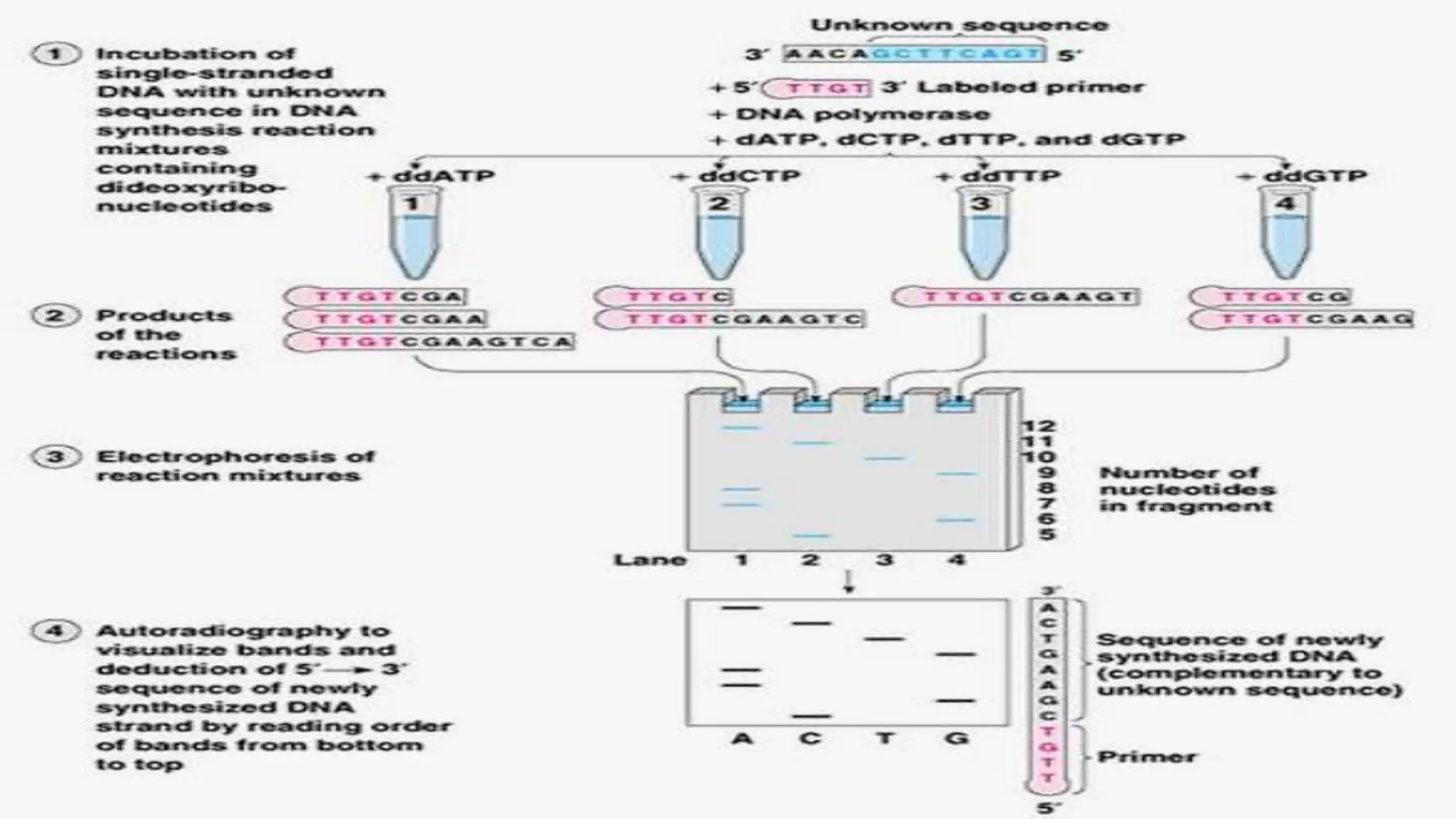
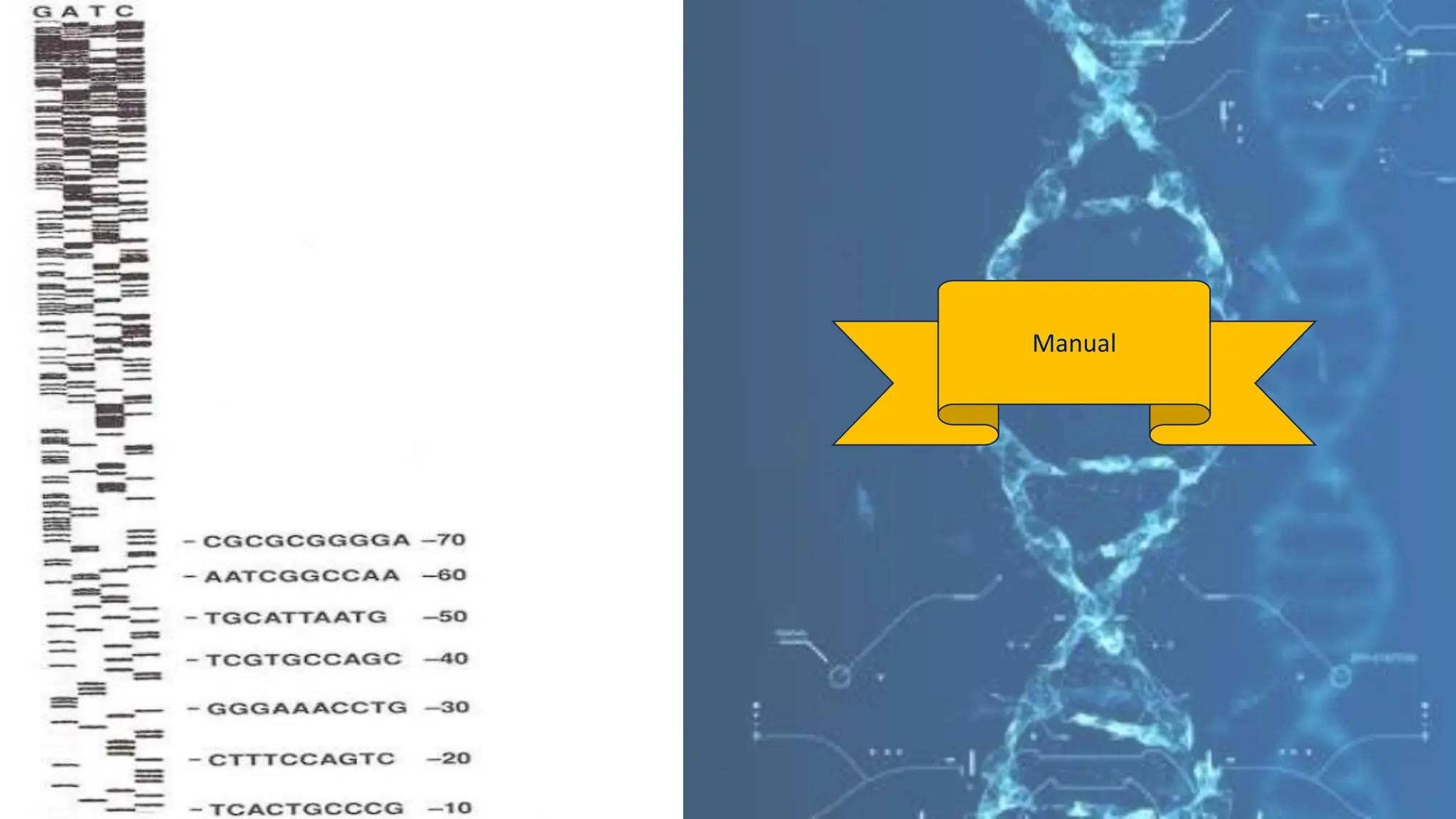
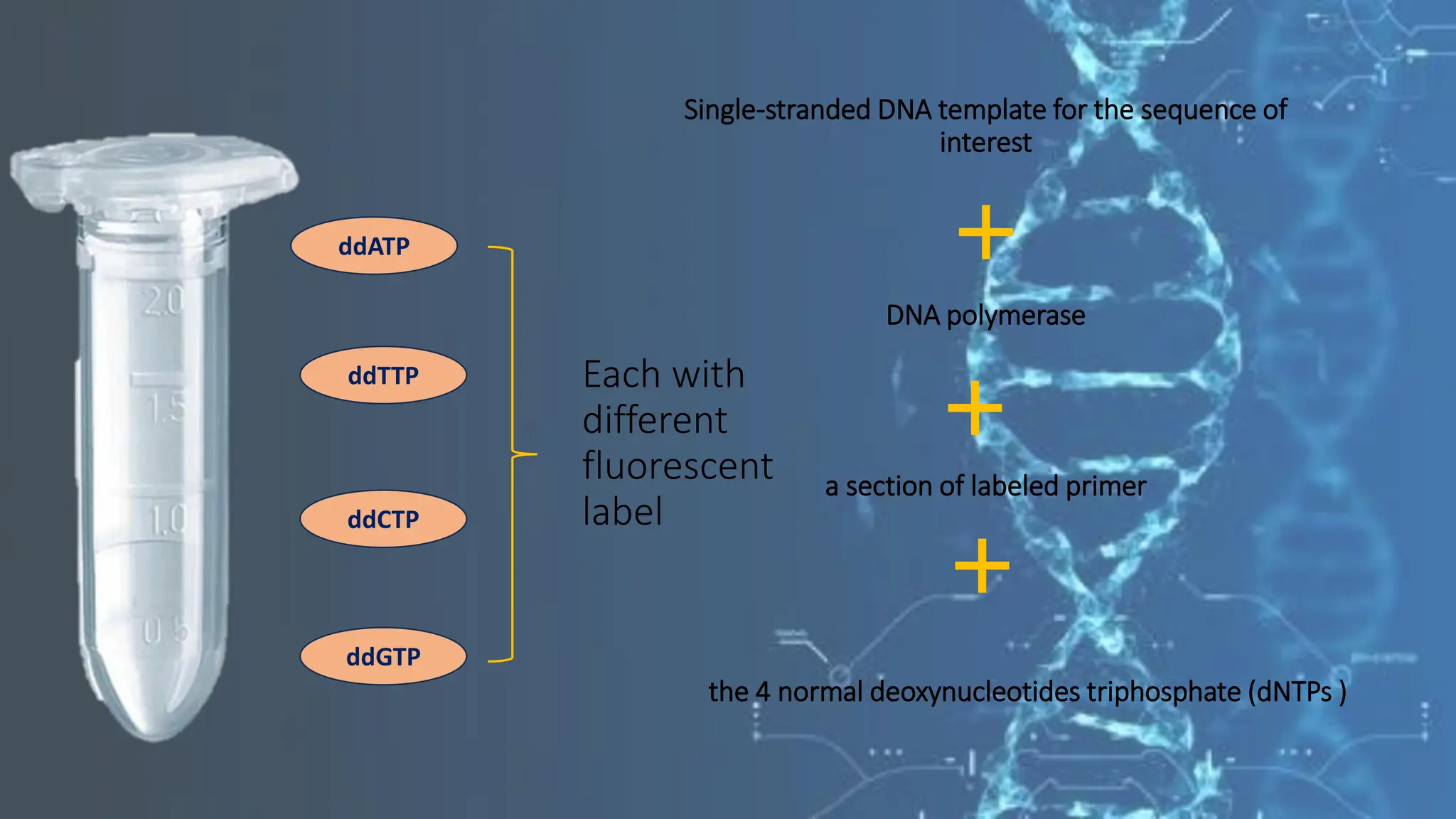
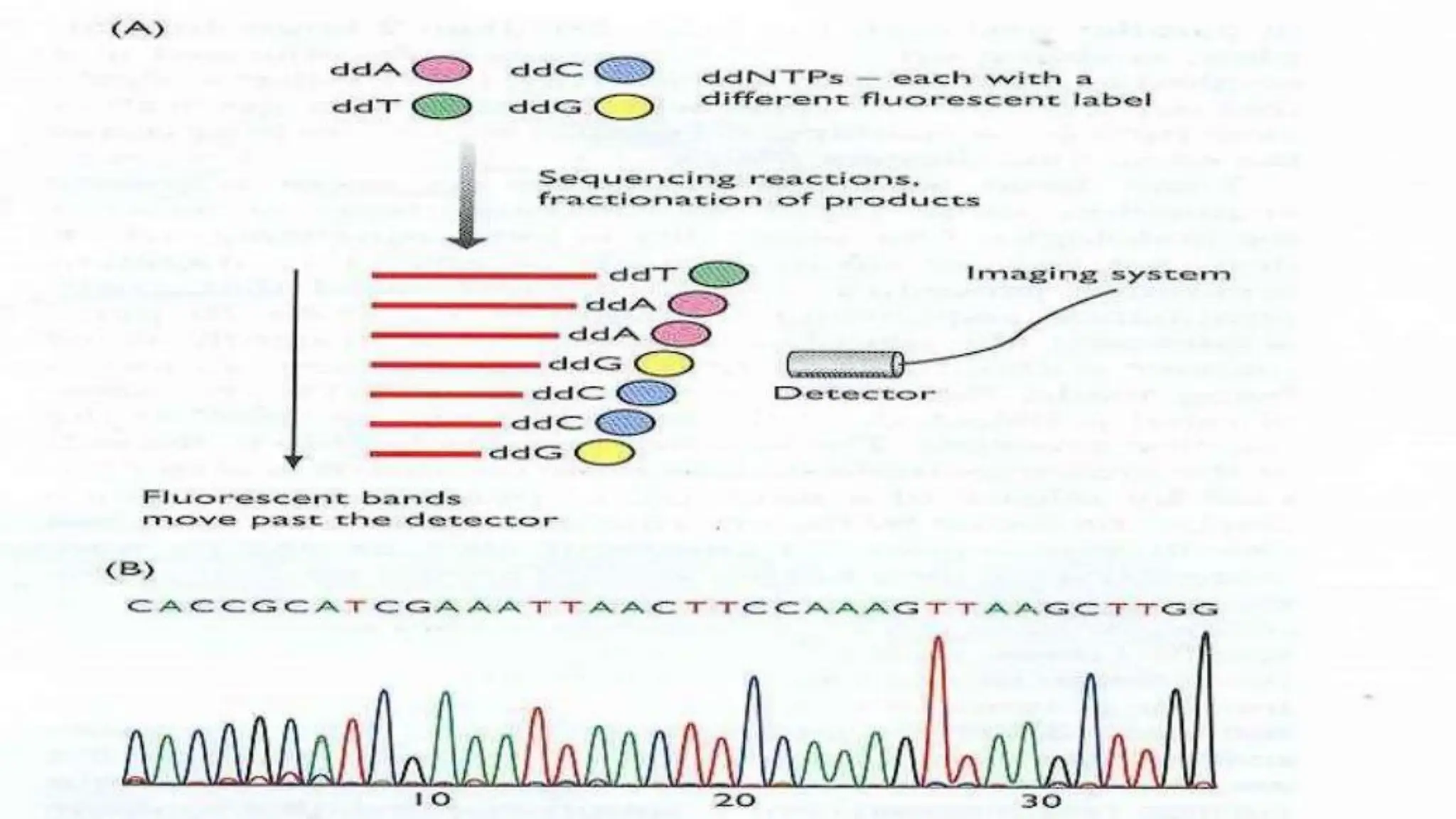
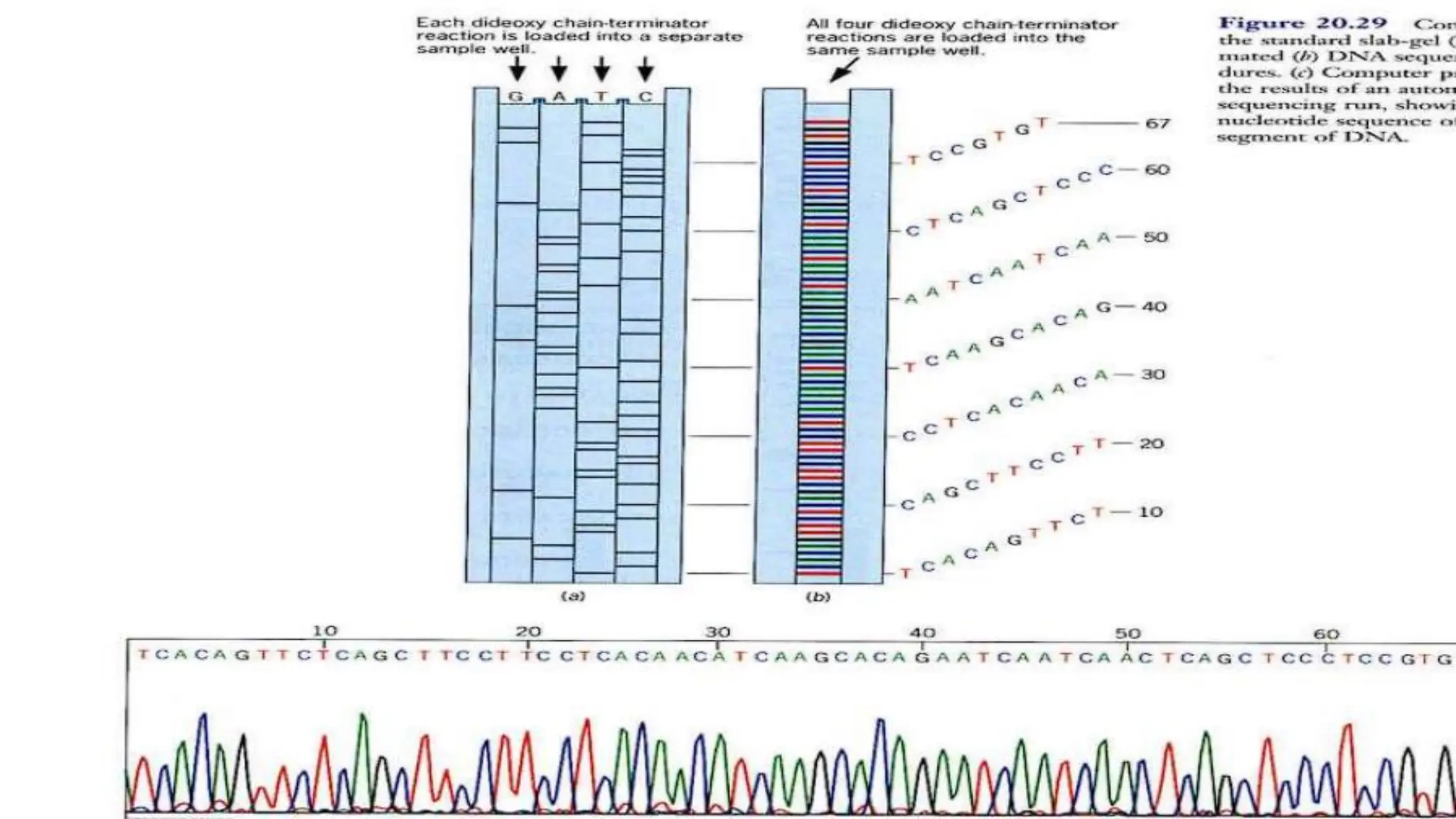
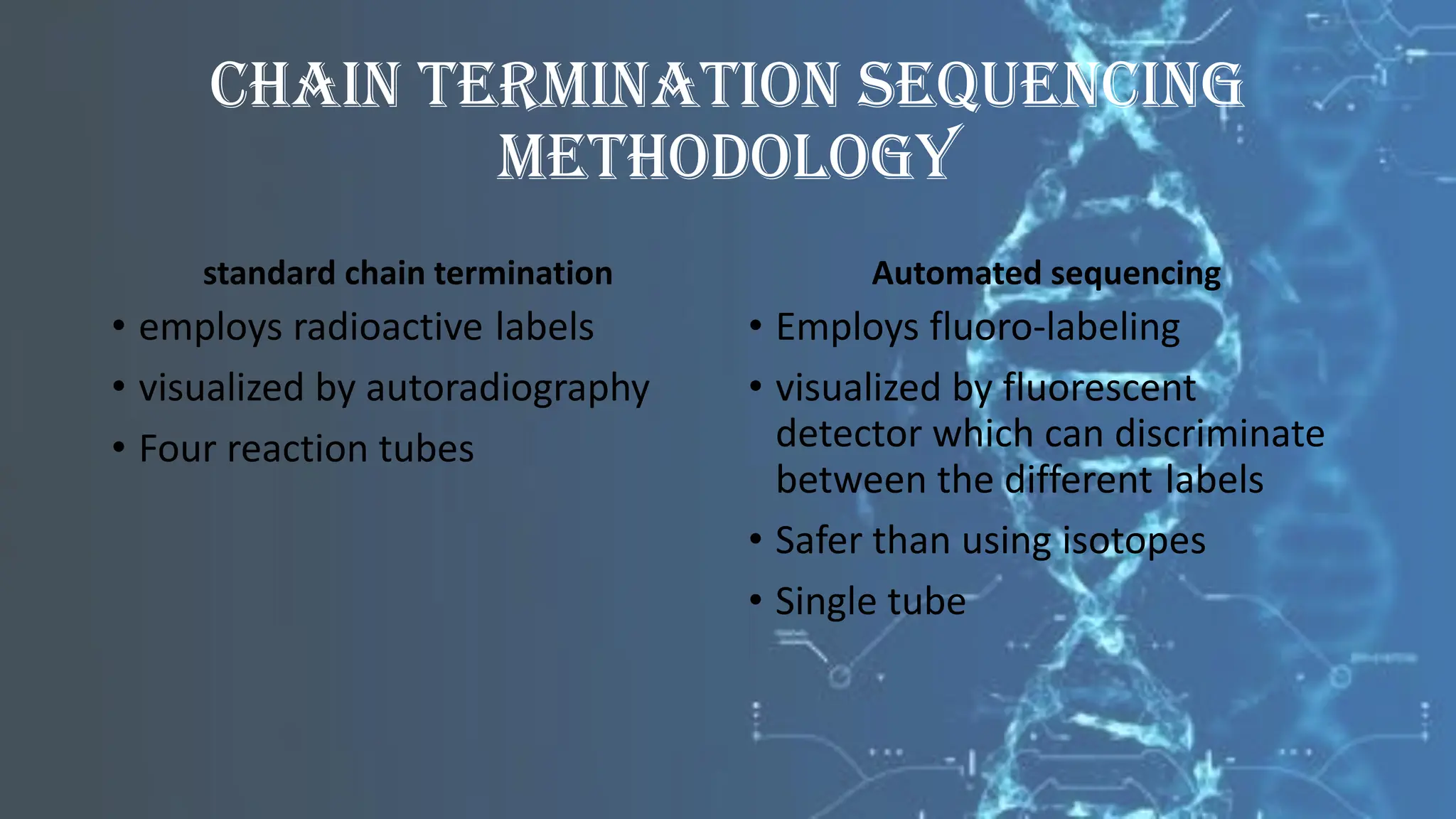
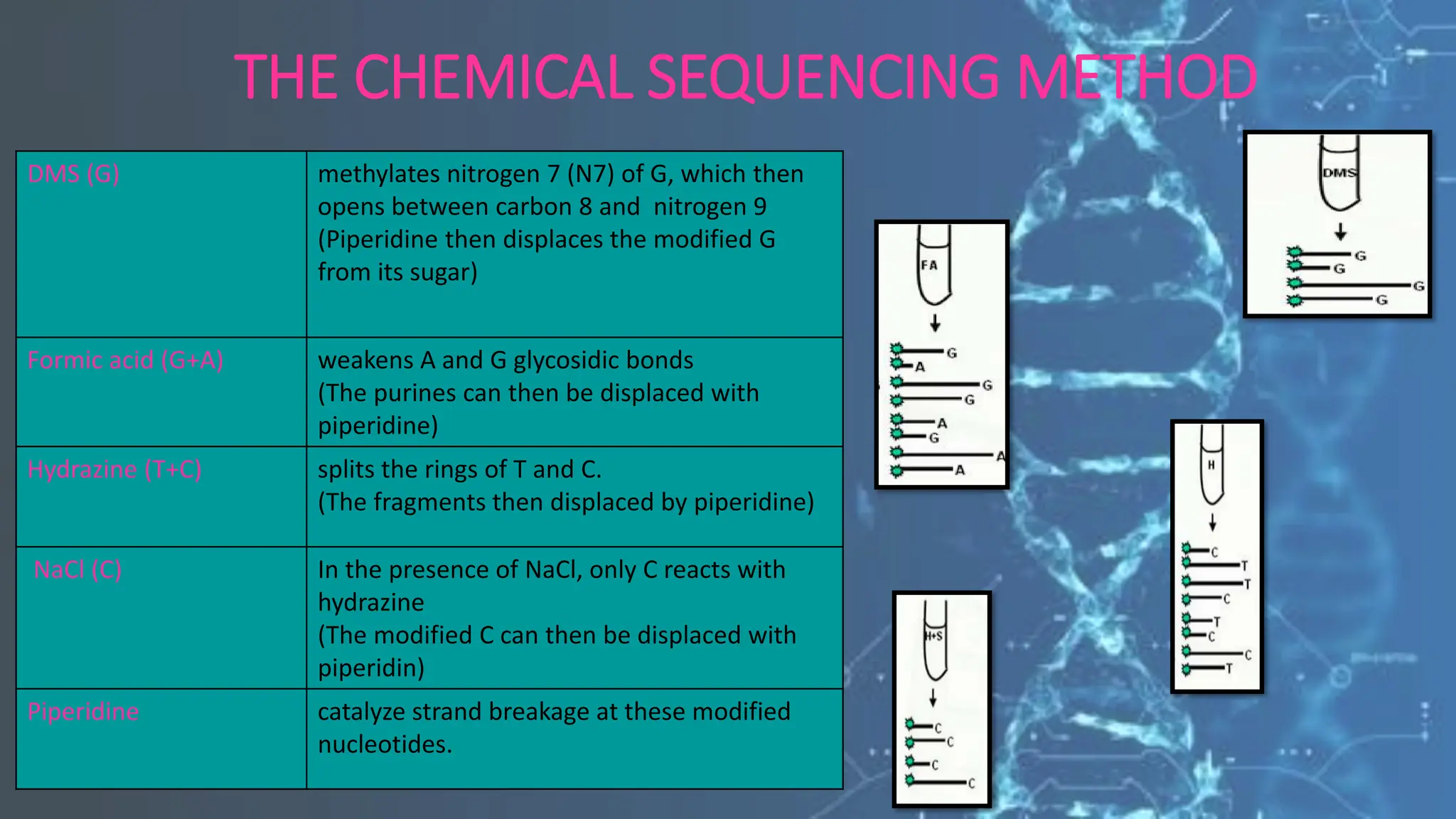
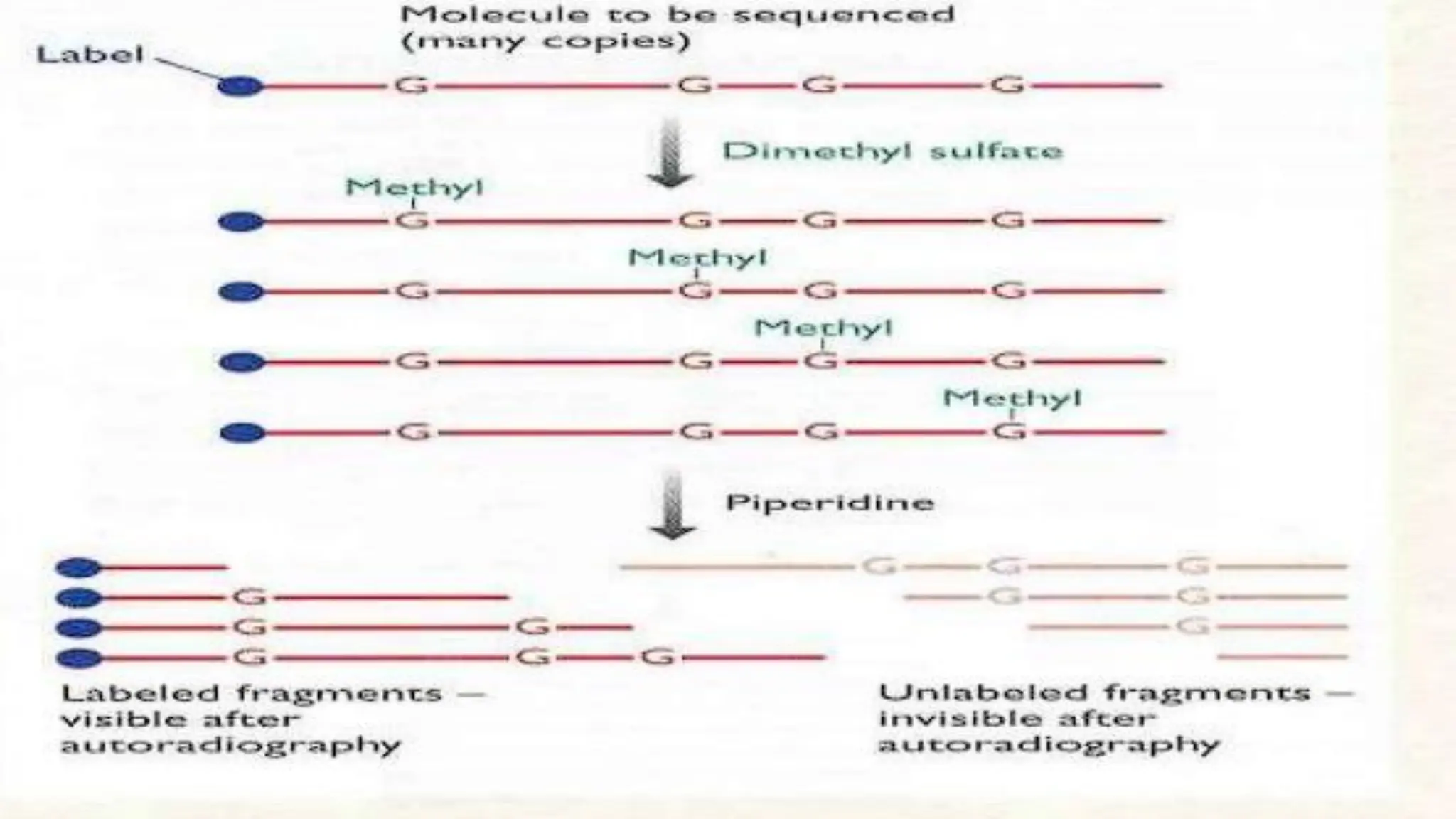
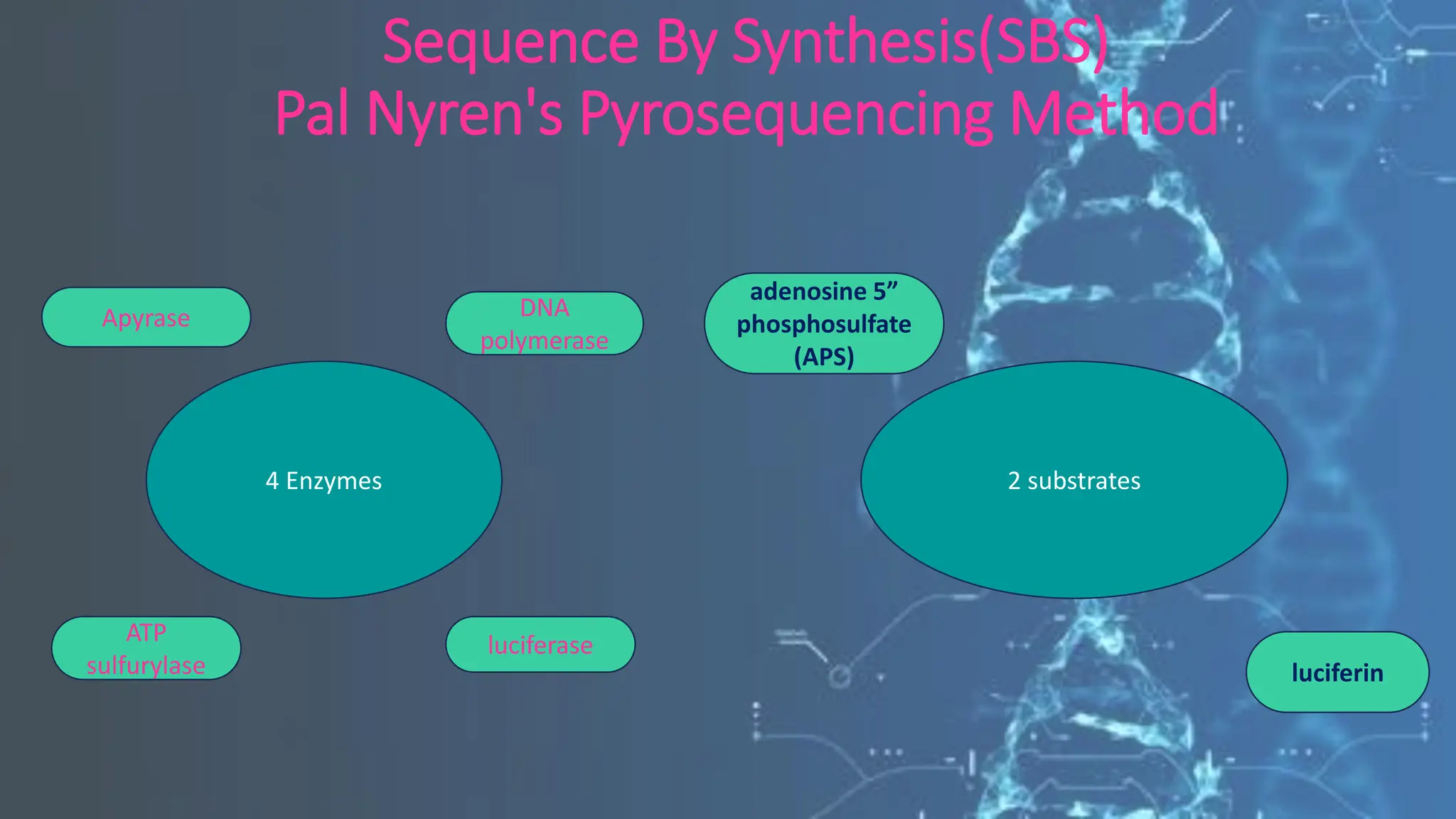
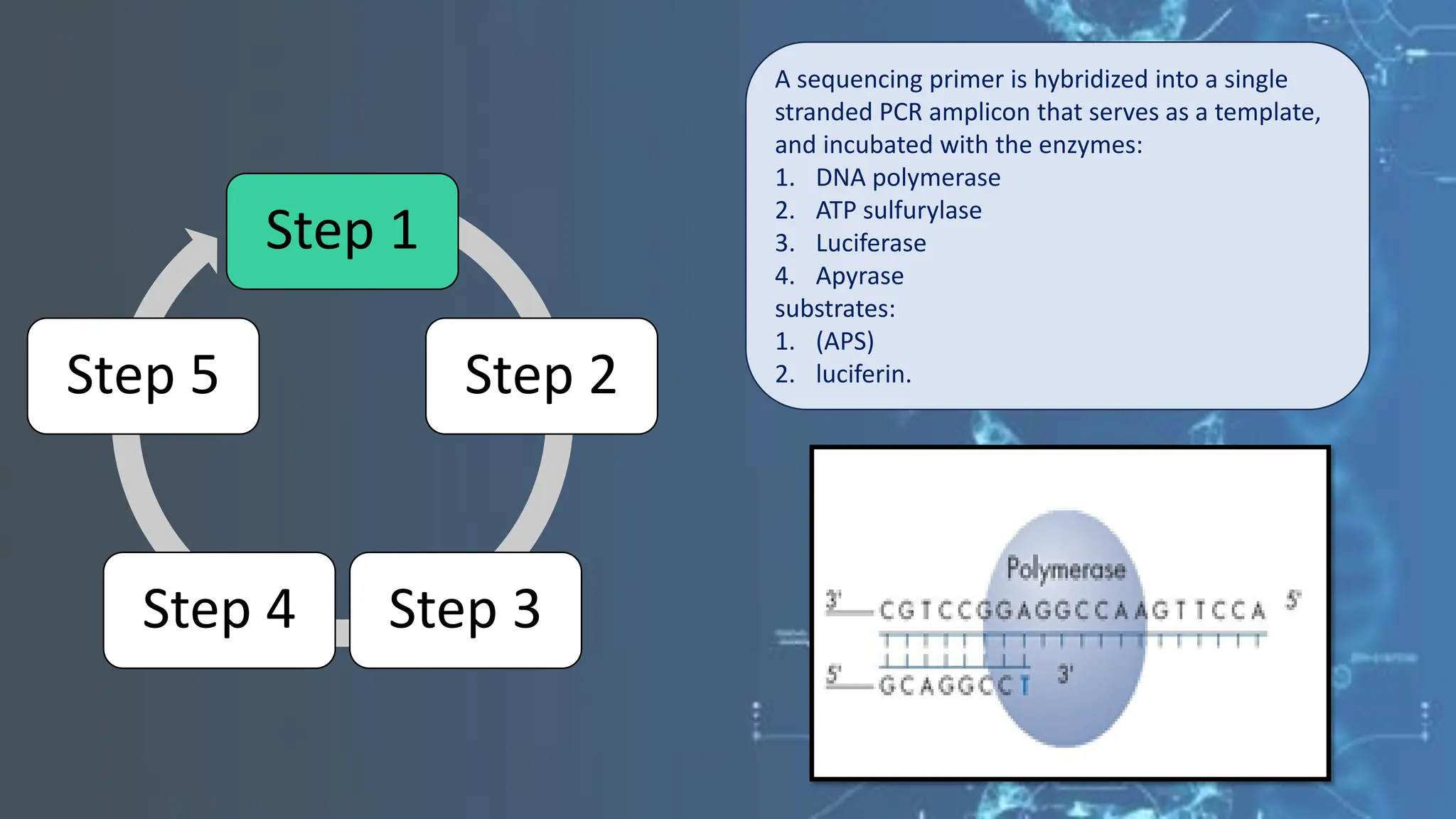
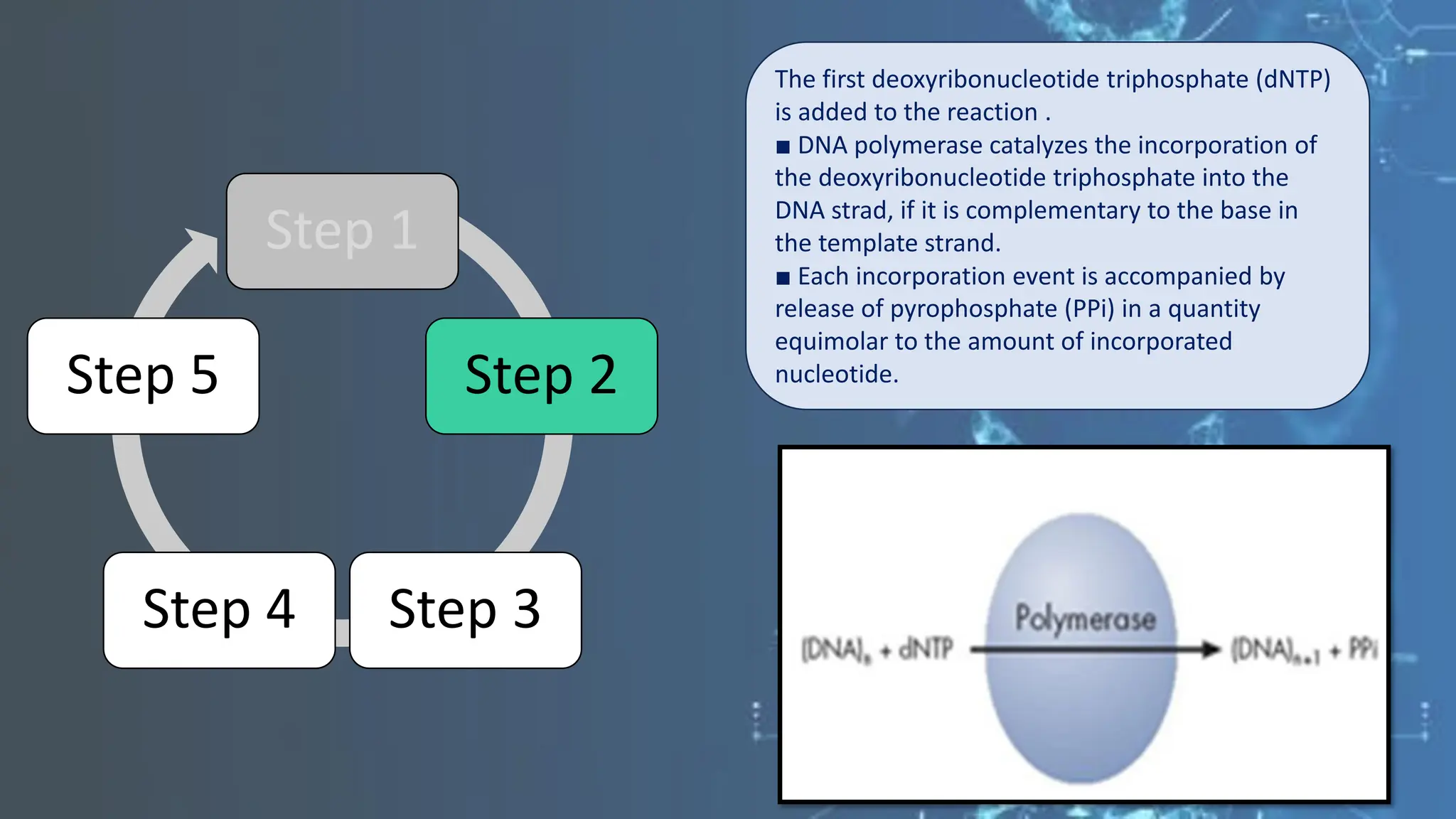
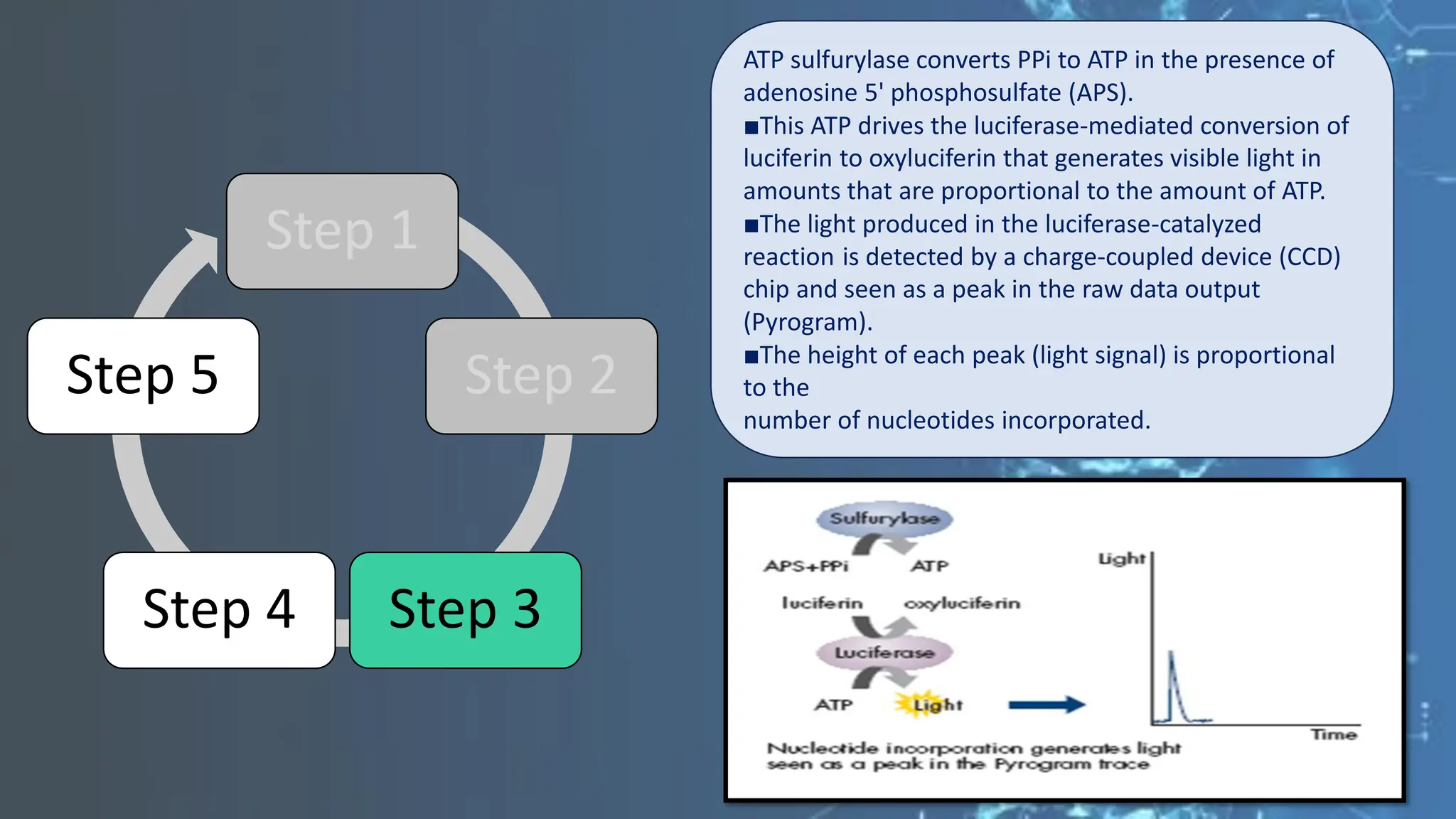
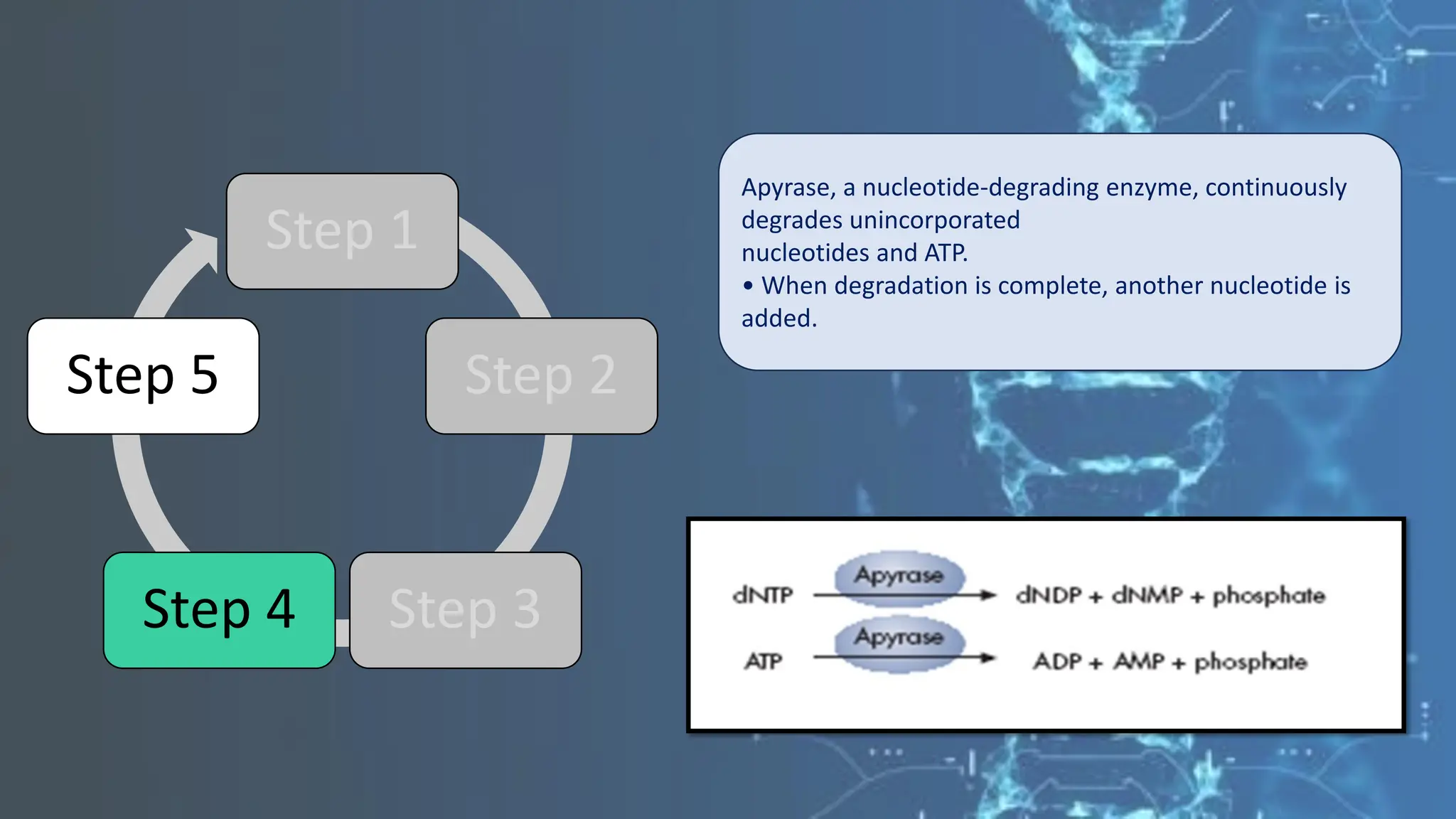
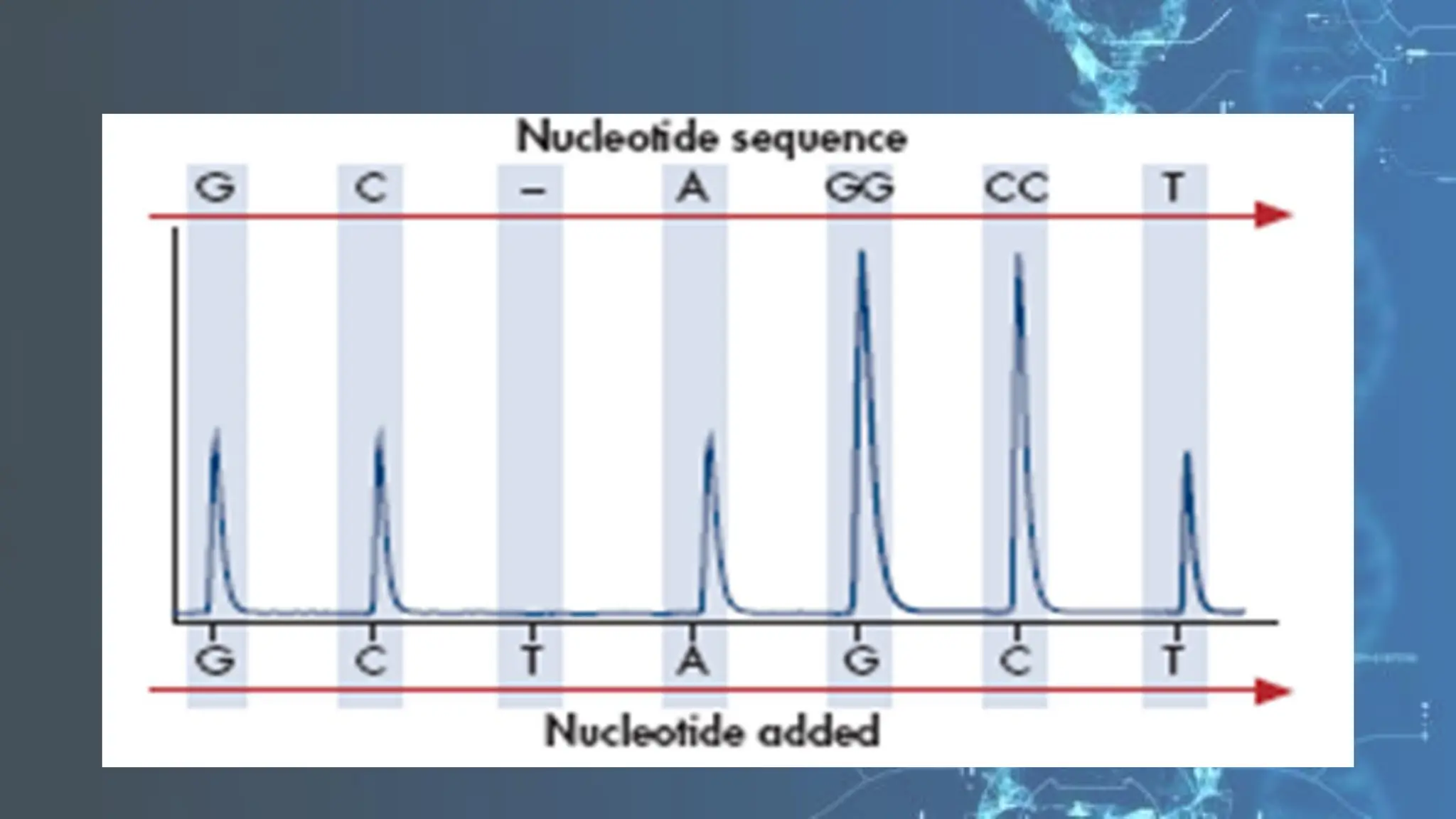
The document details the steps and methodologies for DNA sequencing, emphasizing its significance for genome mapping and manipulation in recombinant DNA experiments. It outlines traditional methods such as Sanger and Maxam-Gilbert sequencing, and introduces next-generation sequencing (NGS) as a transformative technology that allows for efficient and large-scale sequencing. Important mechanisms, including chain termination and sequencing by synthesis, are explained in the context of their application to modern genomic studies.