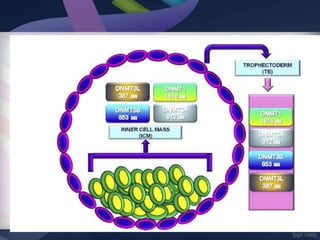
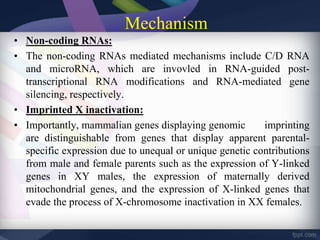
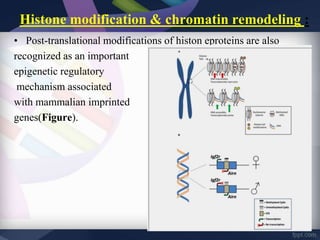
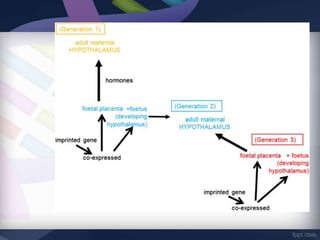
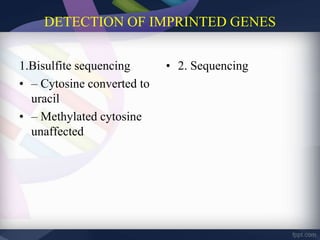
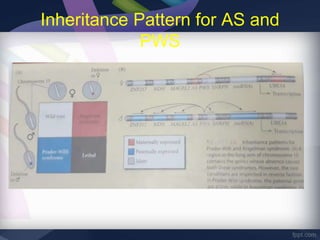
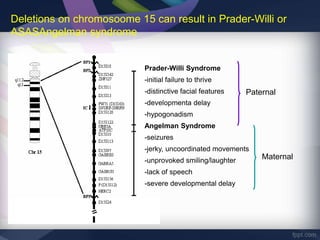
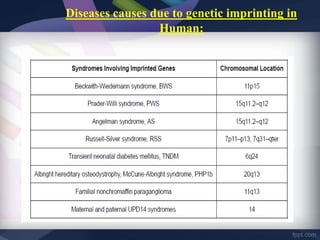
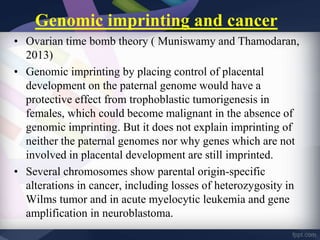
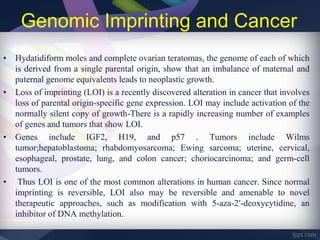
Genomic imprinting refers to genes whose expression depends on whether they are inherited maternally or paternally. Imprinted genes are regulated by epigenetic mechanisms like DNA methylation and histone modifications. Disruption of imprinting can cause diseases in both humans and animals. In humans, imprinting disorders include Prader-Willi and Angelman syndromes, which result from deletions on chromosome 15 and can be paternal or maternal in origin. In animals, disruption of imprinting can cause conditions like large offspring syndrome.