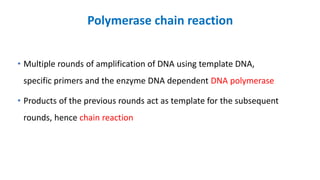
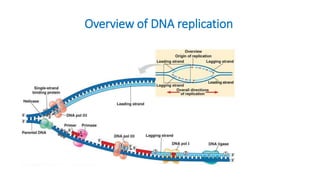
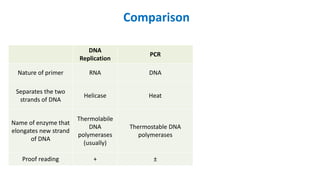
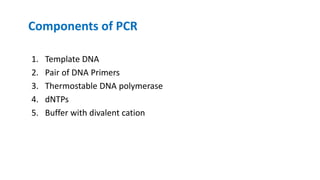
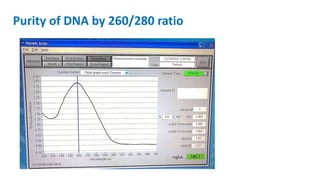
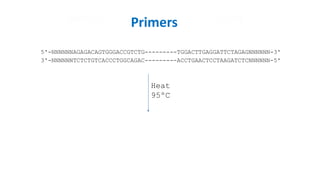
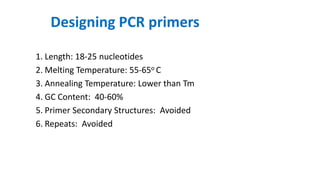
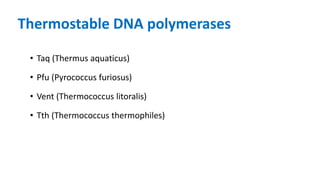
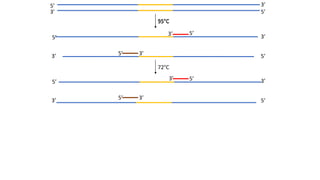
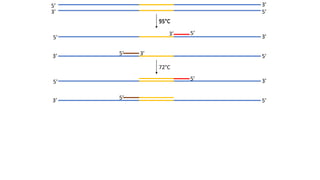
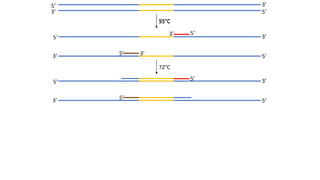
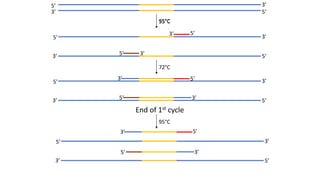
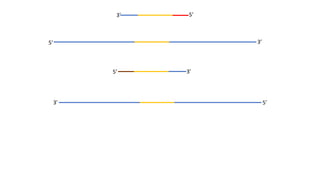
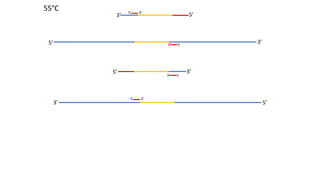
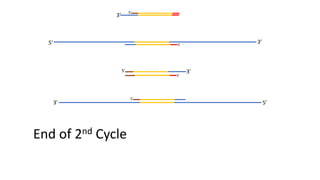
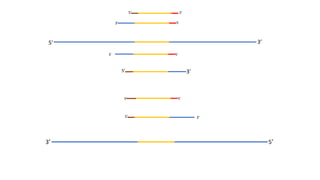
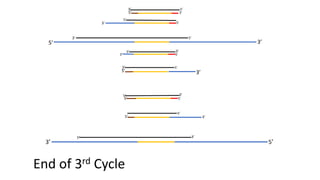
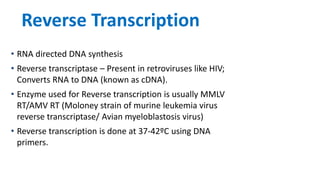
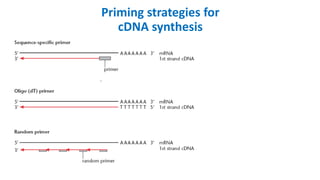
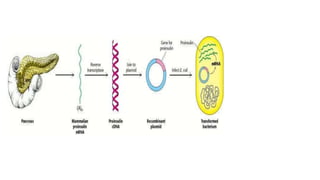
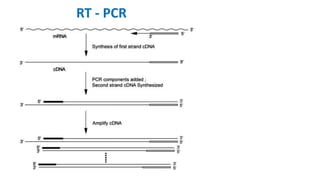
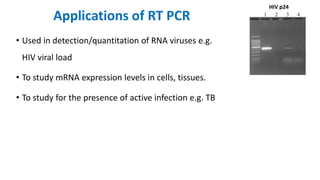
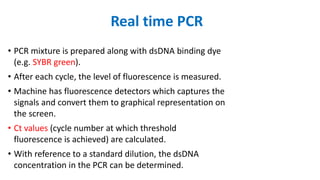
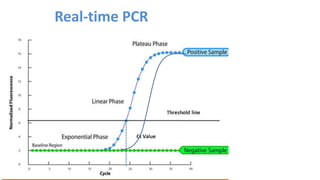
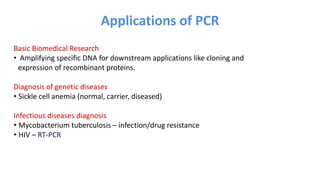
The document outlines the details of the polymerase chain reaction (PCR), an enzymatic DNA replication technique used for amplifying specific DNA segments. It covers the components required for PCR, such as template DNA, primers, and thermostable DNA polymerases, along with the processes involved, including real-time PCR and reverse transcription PCR. The advantages and applications of PCR in various fields like biomedical research, genetic disease diagnosis, and forensics are also discussed.