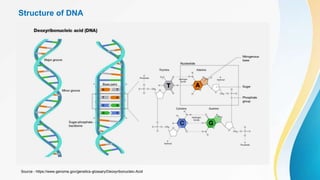
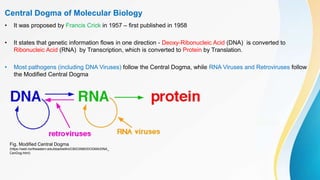
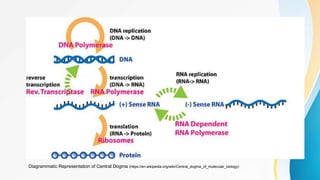
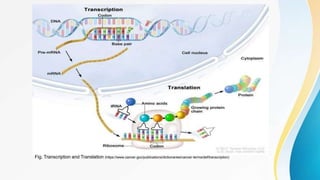
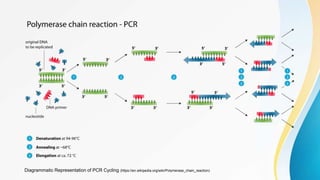
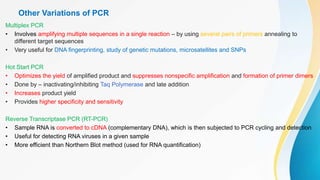
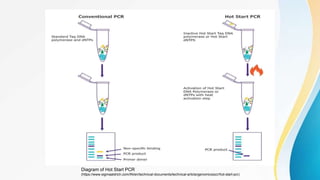
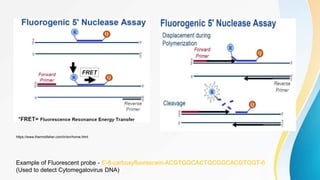
This document provides an overview of central dogma of molecular biology and polymerase chain reaction (PCR). It discusses the structure of DNA and key figures in DNA discovery like Rosalind Franklin. It then explains central dogma, which describes how genetic information flows from DNA to RNA to protein. The document outlines the basic process and components of PCR including DNA template, primers, DNA polymerase, thermal cycler. It also discusses variations like real-time PCR, hot start PCR, and nested PCR. Equipment used in PCR like thermal cyclers, centrifuges and spectrophotometers are also described.