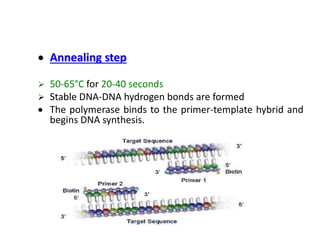
The document discusses polymerase chain reaction (PCR), invented by Dr. Kary Mullis in 1983, which allows for the amplification of specific DNA sequences. It outlines various types of PCR and their applications in fields such as genetic engineering, disease detection, and forensic science, along with detailed steps involved in the PCR process. Additionally, it addresses challenges associated with PCR, such as polymerase errors and size limitations.