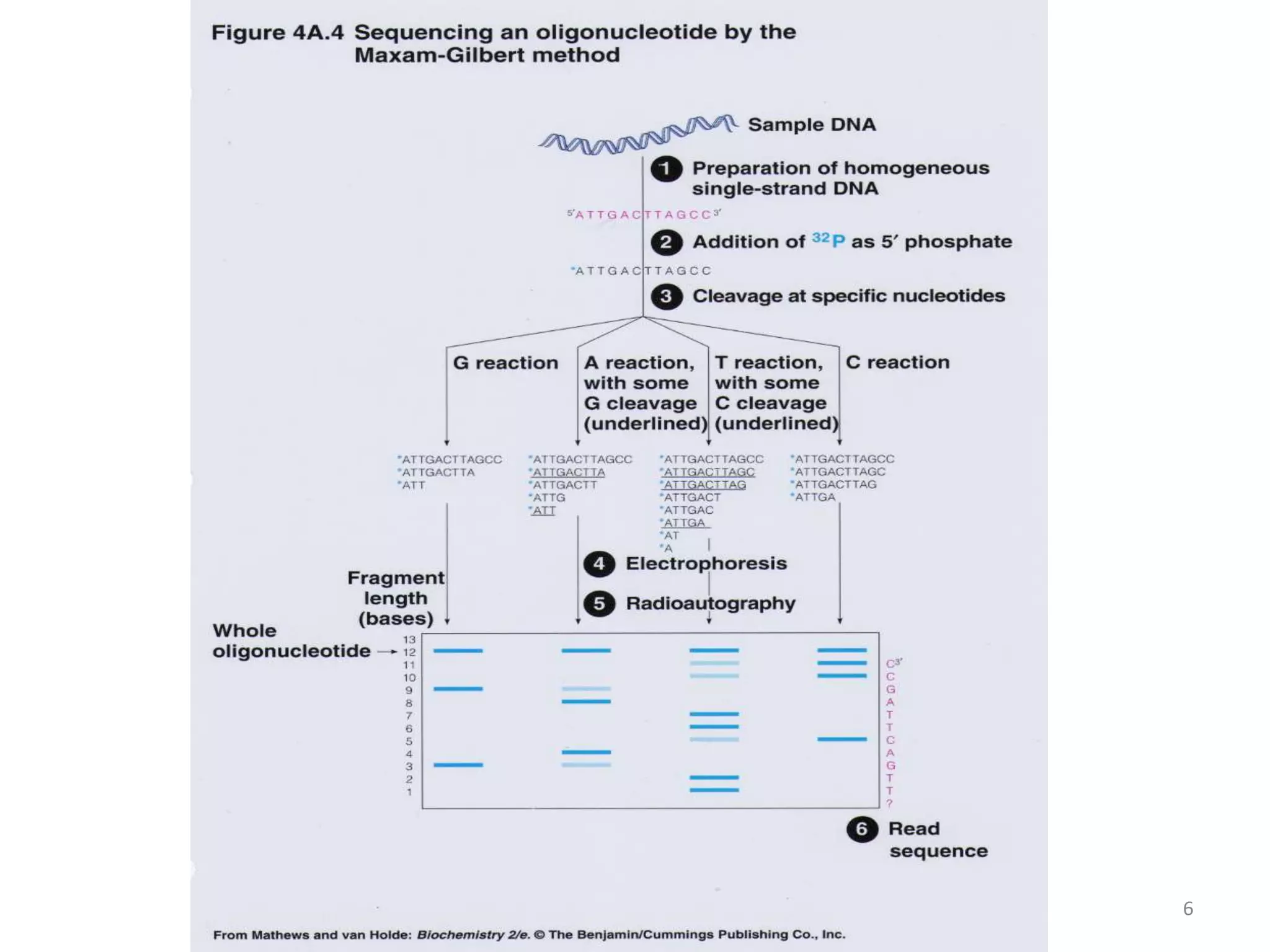
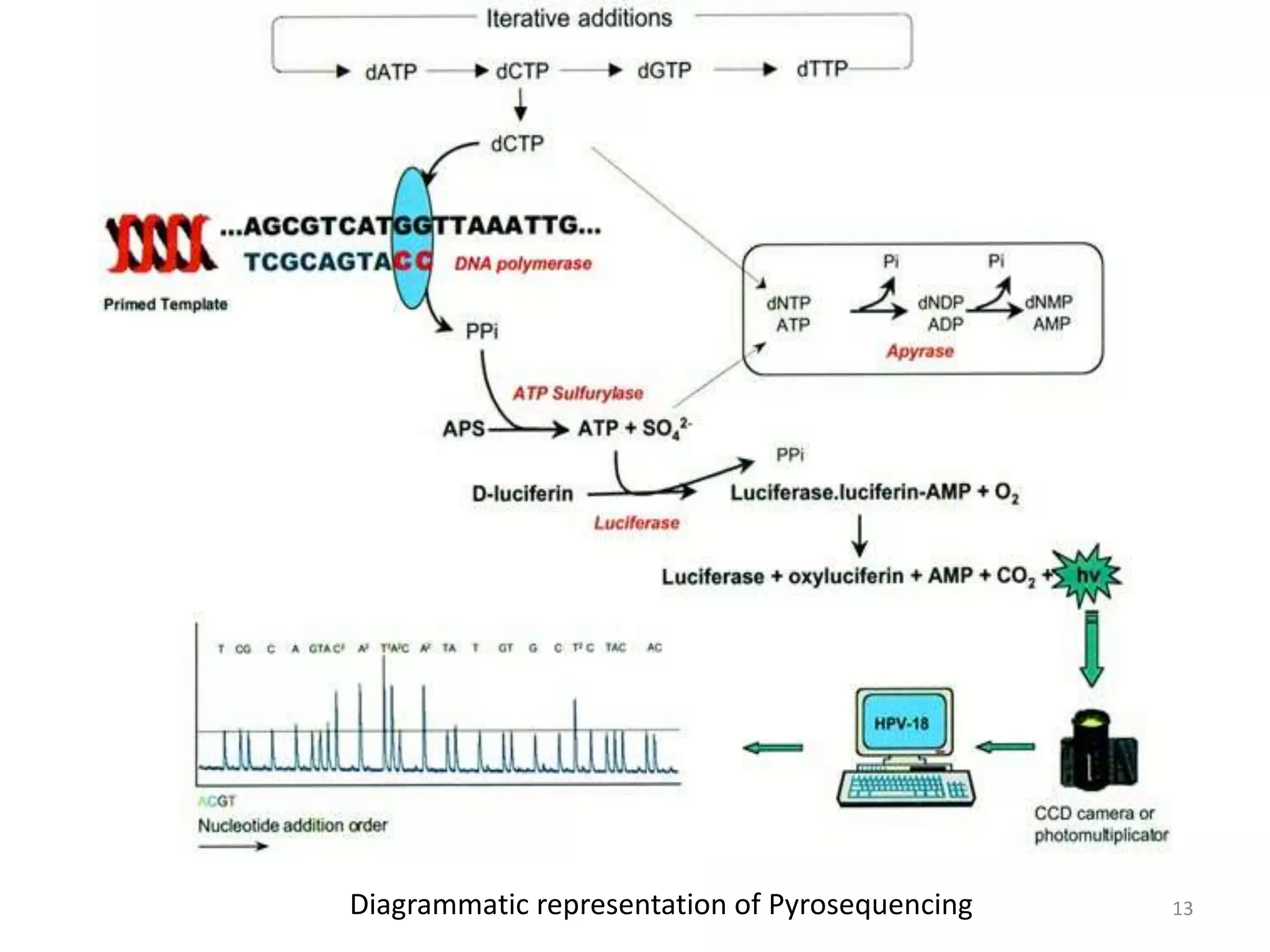
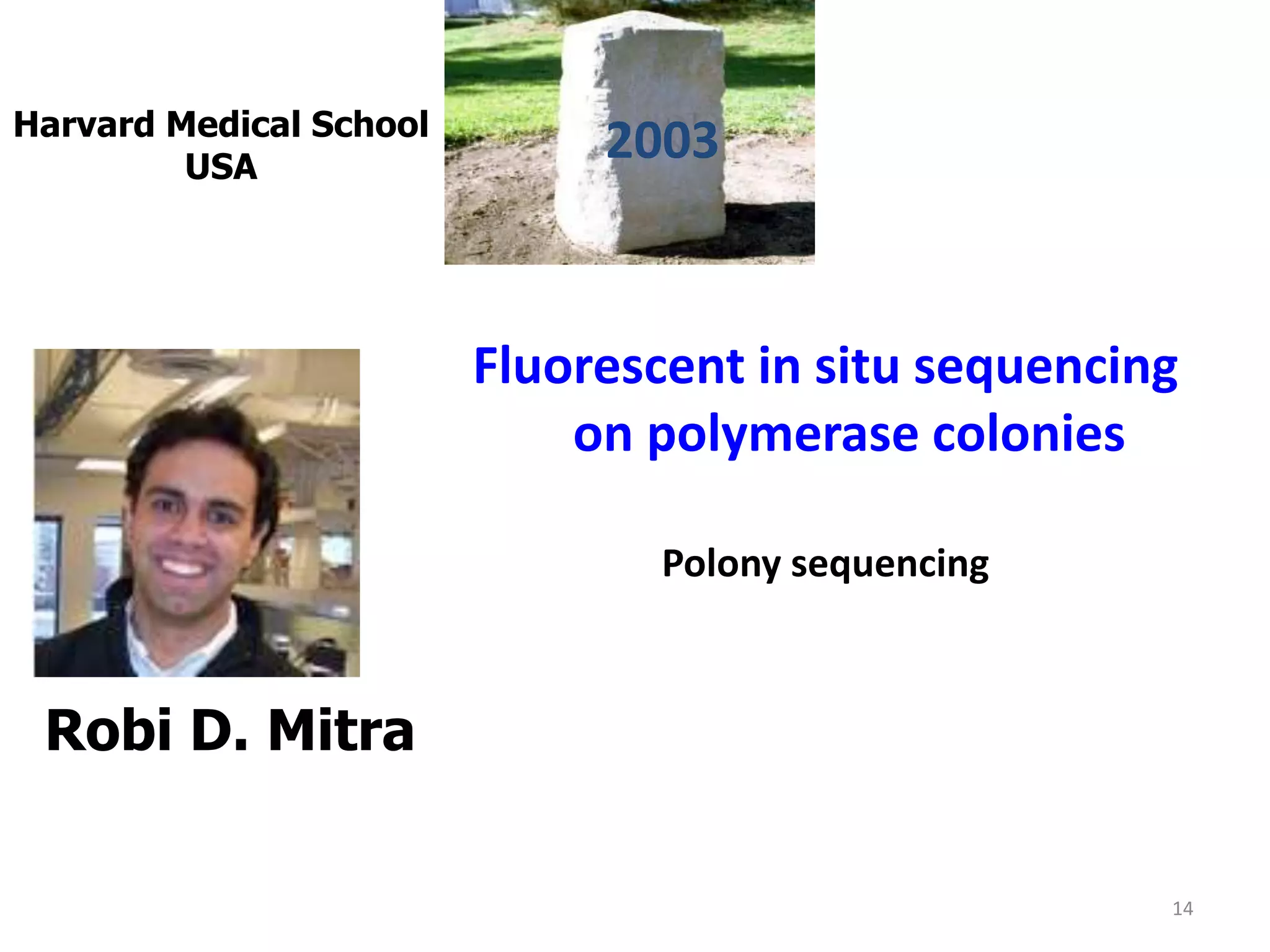
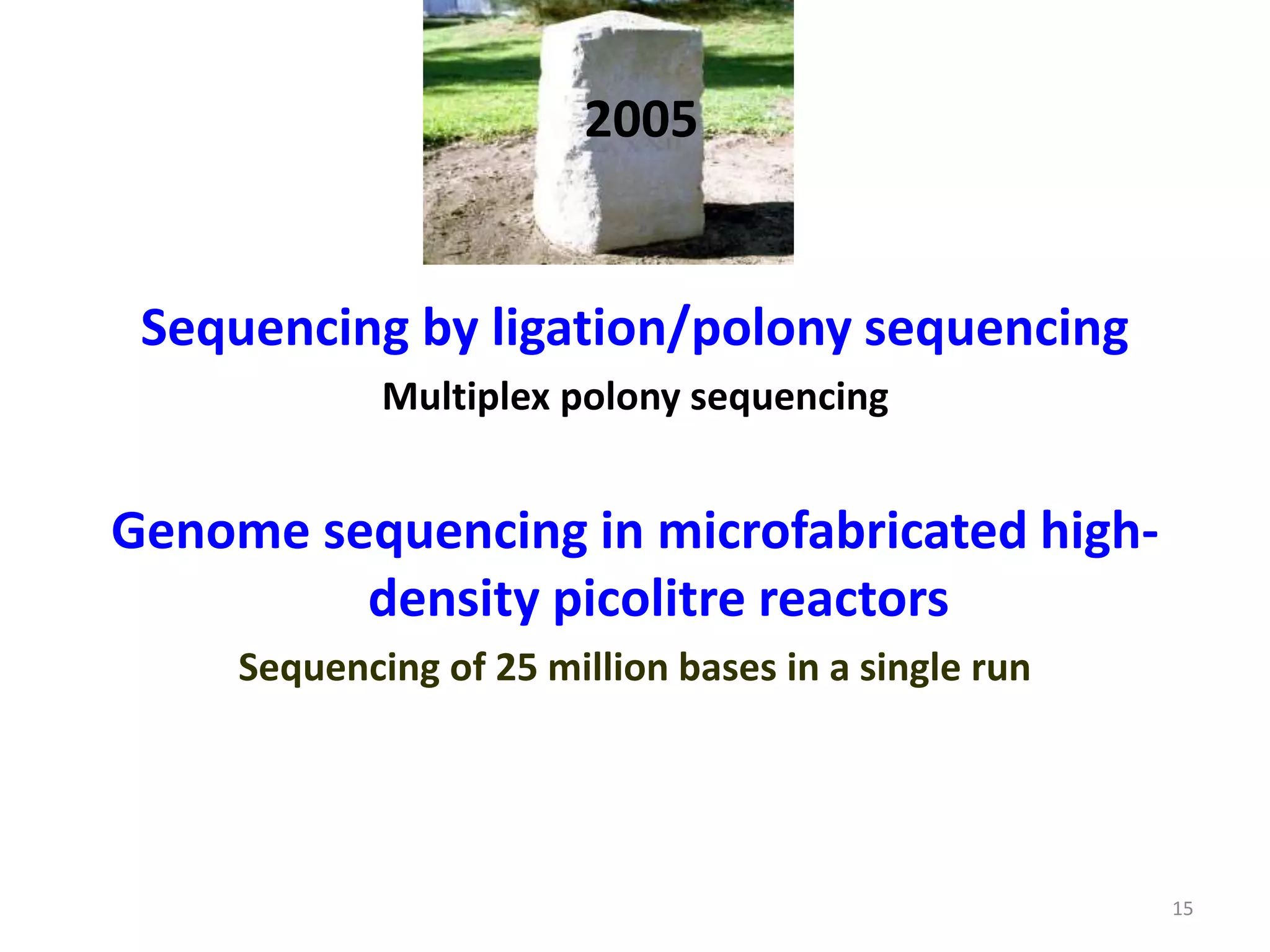
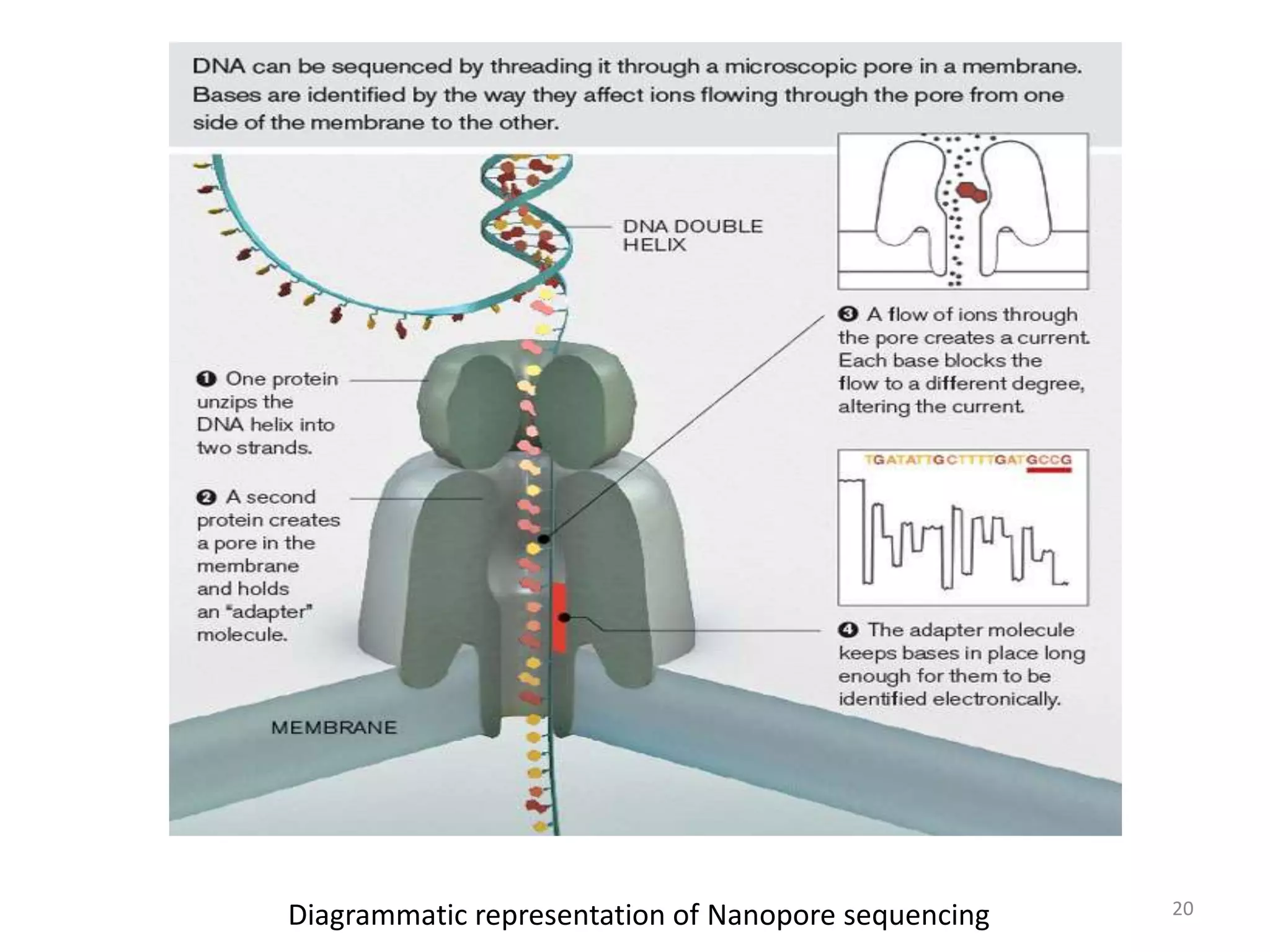
The document describes several methods for DNA sequencing, including Maxam-Gilbert chemical degradation, pyrosequencing, polony sequencing, and nanopore sequencing. The Maxam-Gilbert method uses chemical reactions to cleave DNA at specific bases, followed by gel electrophoresis. Pyrosequencing sequences DNA by detecting pyrophosphate release during nucleotide incorporation. Polony sequencing performs sequencing on polymerase colonies using fluorescent in situ hybridization. Nanopore sequencing detects DNA sequences as single strands pass through nanopores.