The document provides information on several DNA sequencing methods including:
- The Maxam-Gilbert method which uses chemical cleavage to sequence DNA.
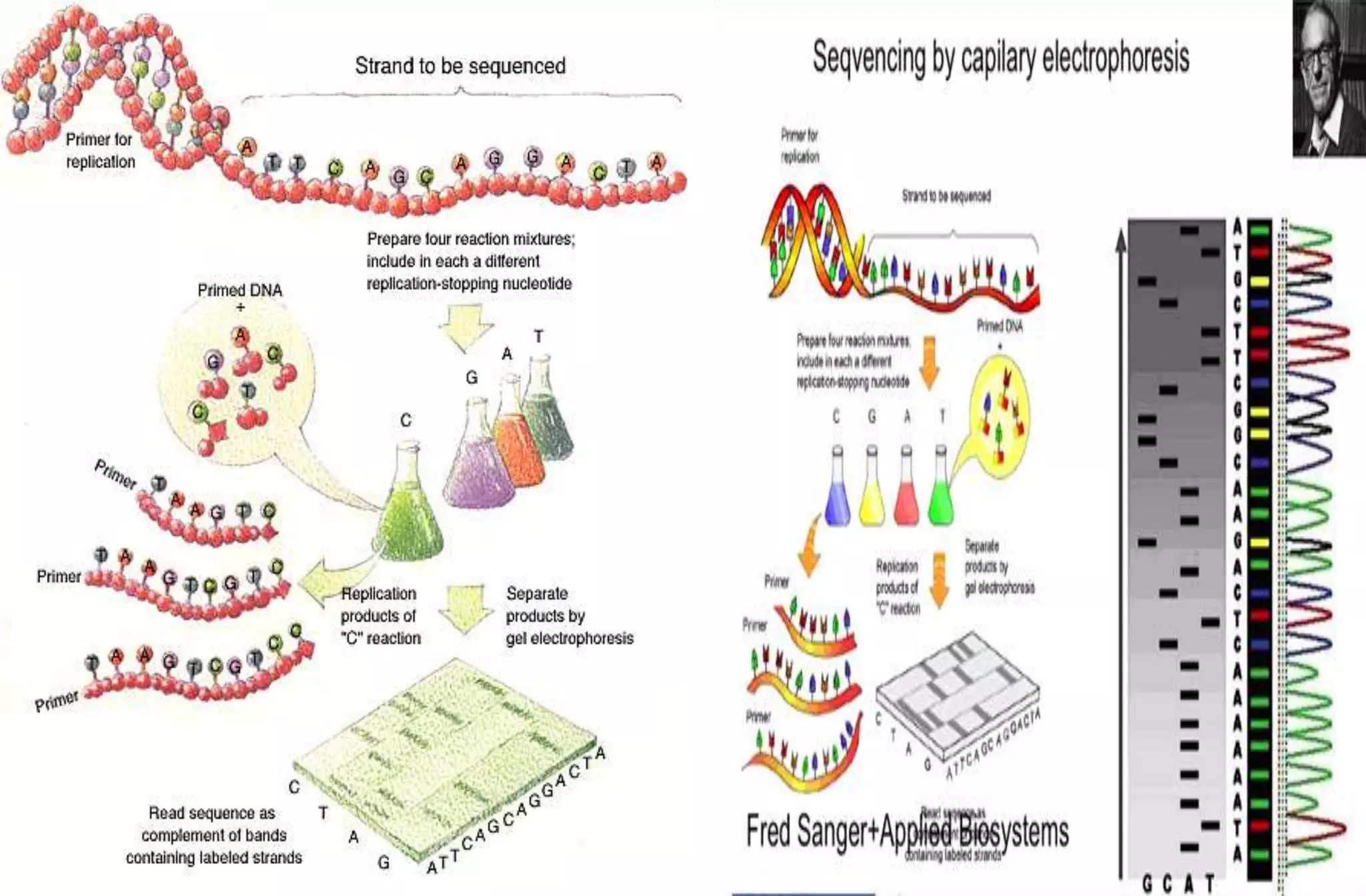
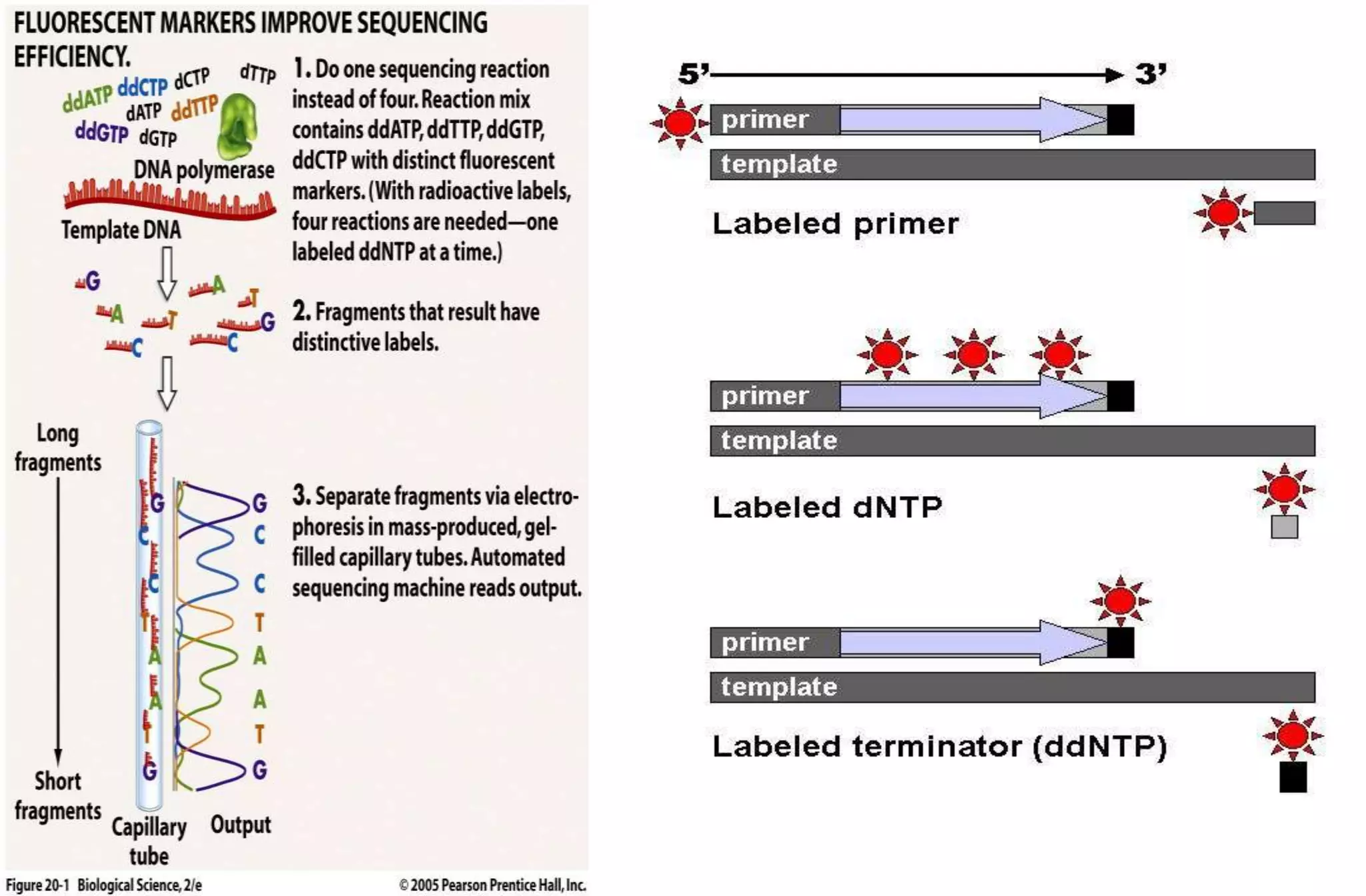
- The Sanger method (chain-termination method) which uses dideoxynucleotides and DNA polymerase to terminate DNA strand extension at specific bases.
- Next generation sequencing methods like 454 pyrosequencing, Illumina sequencing, and Ion Torrent sequencing that allow for massively parallel sequencing of many DNA fragments simultaneously.
The history and basic principles of the Maxam-Gilbert and Sanger methods are described in detail as they were the first widely used DNA sequencing techniques. Subsequent methods aimed to improve speed, throughput and reduce cost of sequencing