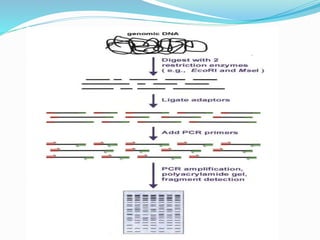
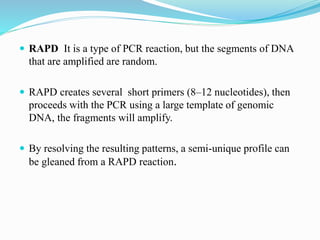
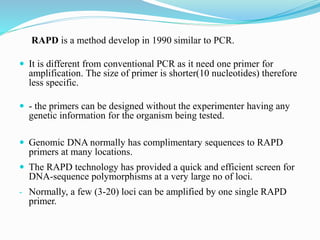
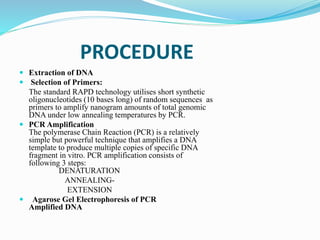
The document provides an overview of various DNA amplification techniques, focusing on Amplified Fragment Length Polymorphism (AFLP) and Randomly Amplified Polymorphic DNA (RAPD). AFLP involves selectively amplifying DNA fragments using two restriction enzymes, yielding high reproducibility and polymorphism, while RAPD uses a single arbitrary primer to amplify DNA and is cost-effective but has limitations in distinguishing genetic variations. Additionally, the document briefly discusses Restriction Fragment Length Polymorphism (RFLP) and Southern blotting as methods for DNA analysis and visualization.