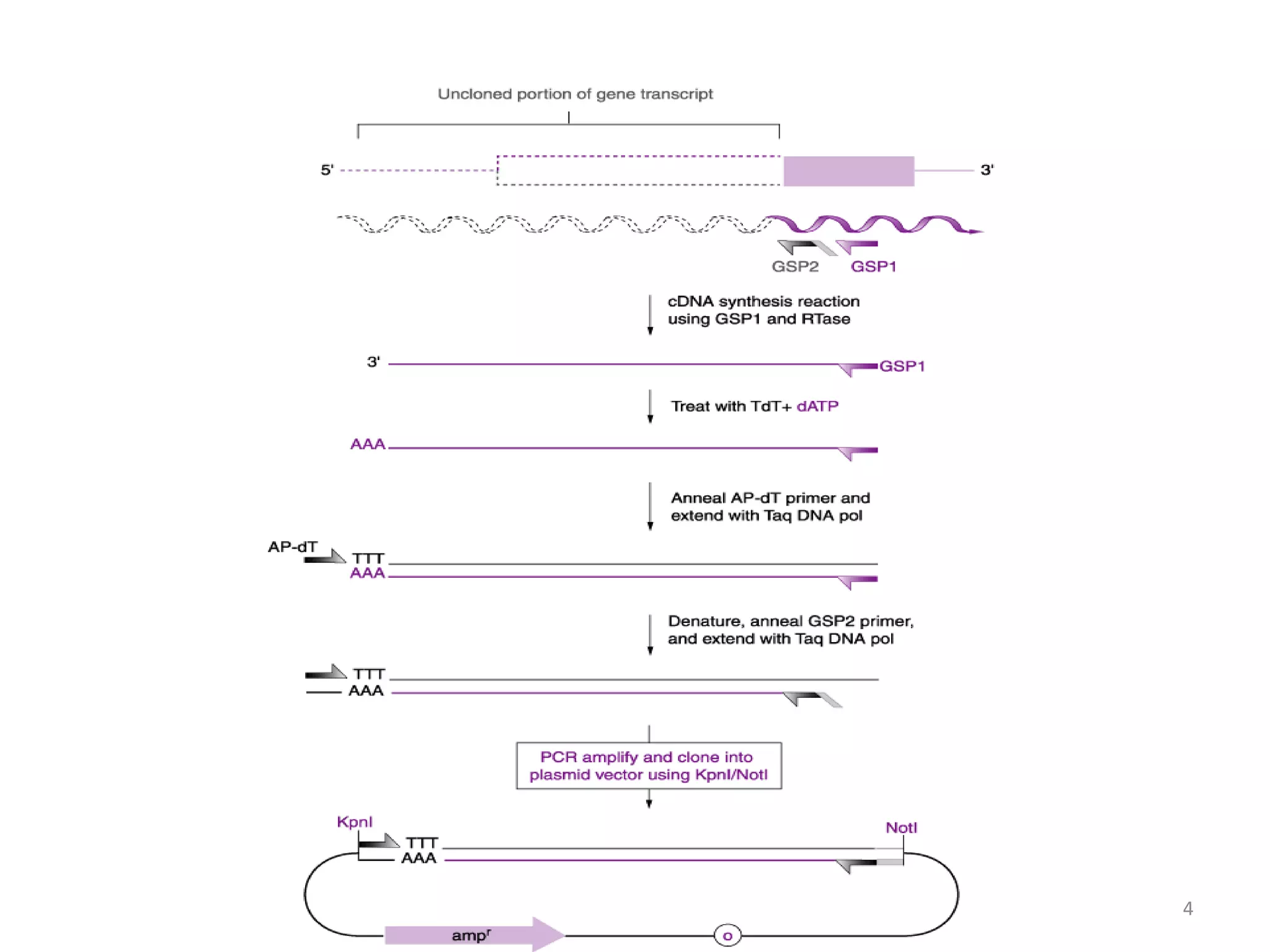
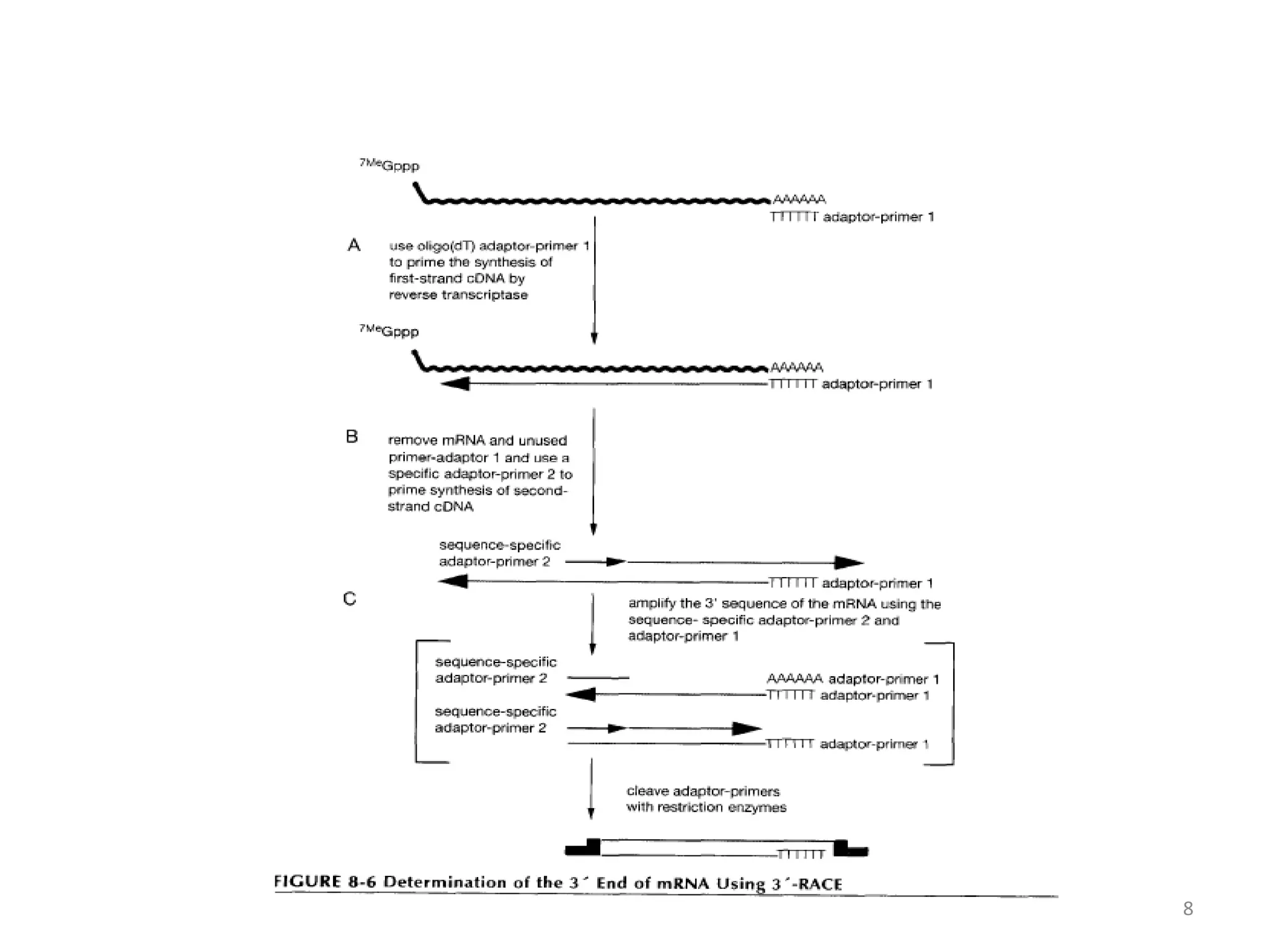
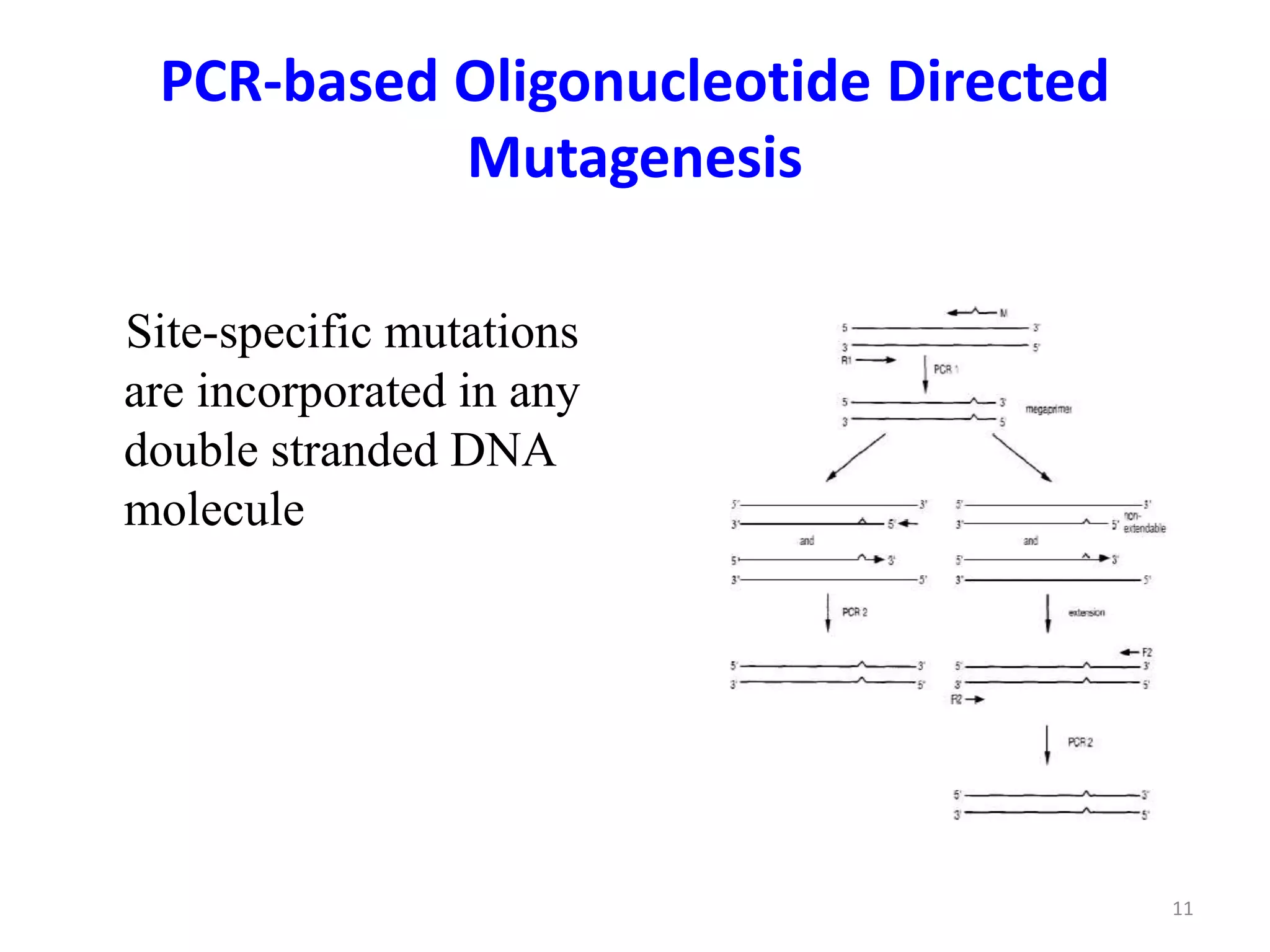
The document discusses various applications of polymerase chain reaction (PCR) including cDNA synthesis and rapid amplification of cDNA ends (RACE) to sequence mRNA ends, error-prone PCR for random mutagenesis, PCR-based molecular markers for genome mapping and analyzing genetic variation, analysis of fossil DNA and environments using PCR, medical diagnosis using PCR, and applications in forensic science.