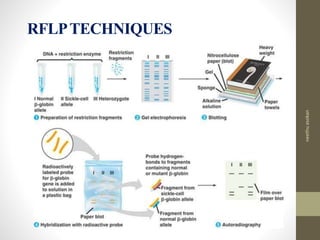
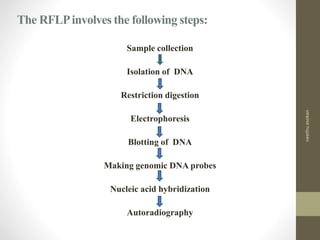
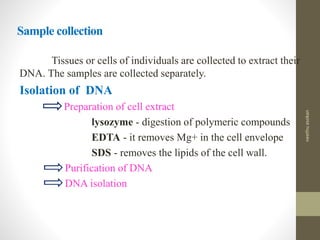
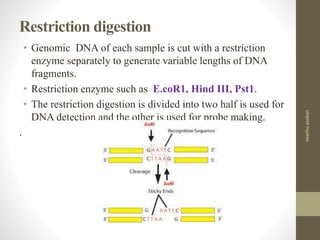
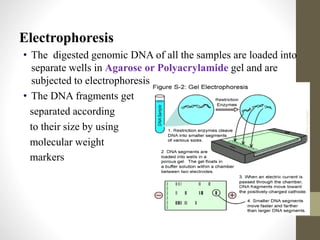
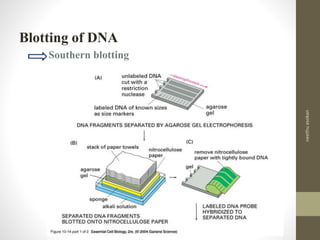
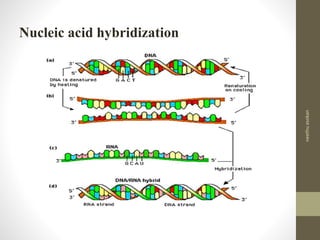
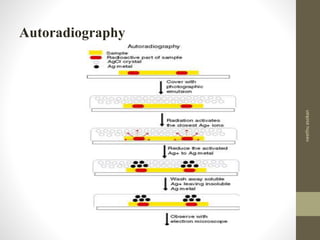
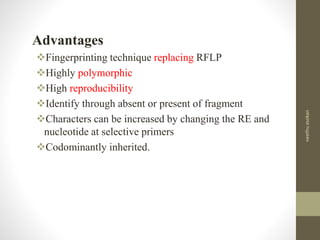
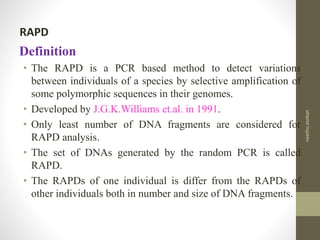
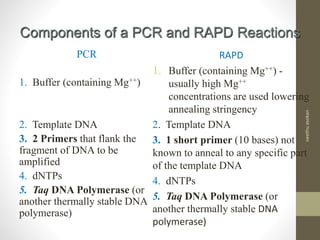
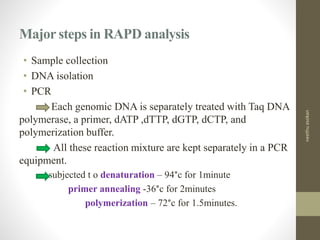
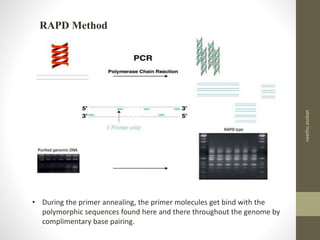
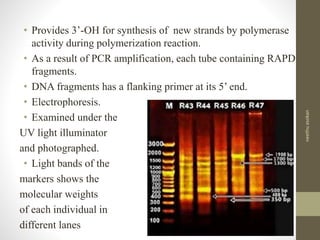
RFLP and RAPD are PCR-based techniques used to analyze genetic variations between individuals. RFLP involves restricting genomic DNA with enzymes, separating fragments via electrophoresis, and comparing patterns. Variations in fragment lengths indicate polymorphisms. RAPD uses short, arbitrary primers to randomly amplify genomic DNA and compare patterns between individuals. Both techniques are useful for constructing genetic maps, identifying genes, distinguishing individuals, and studying genetic diversity and relationships between organisms.