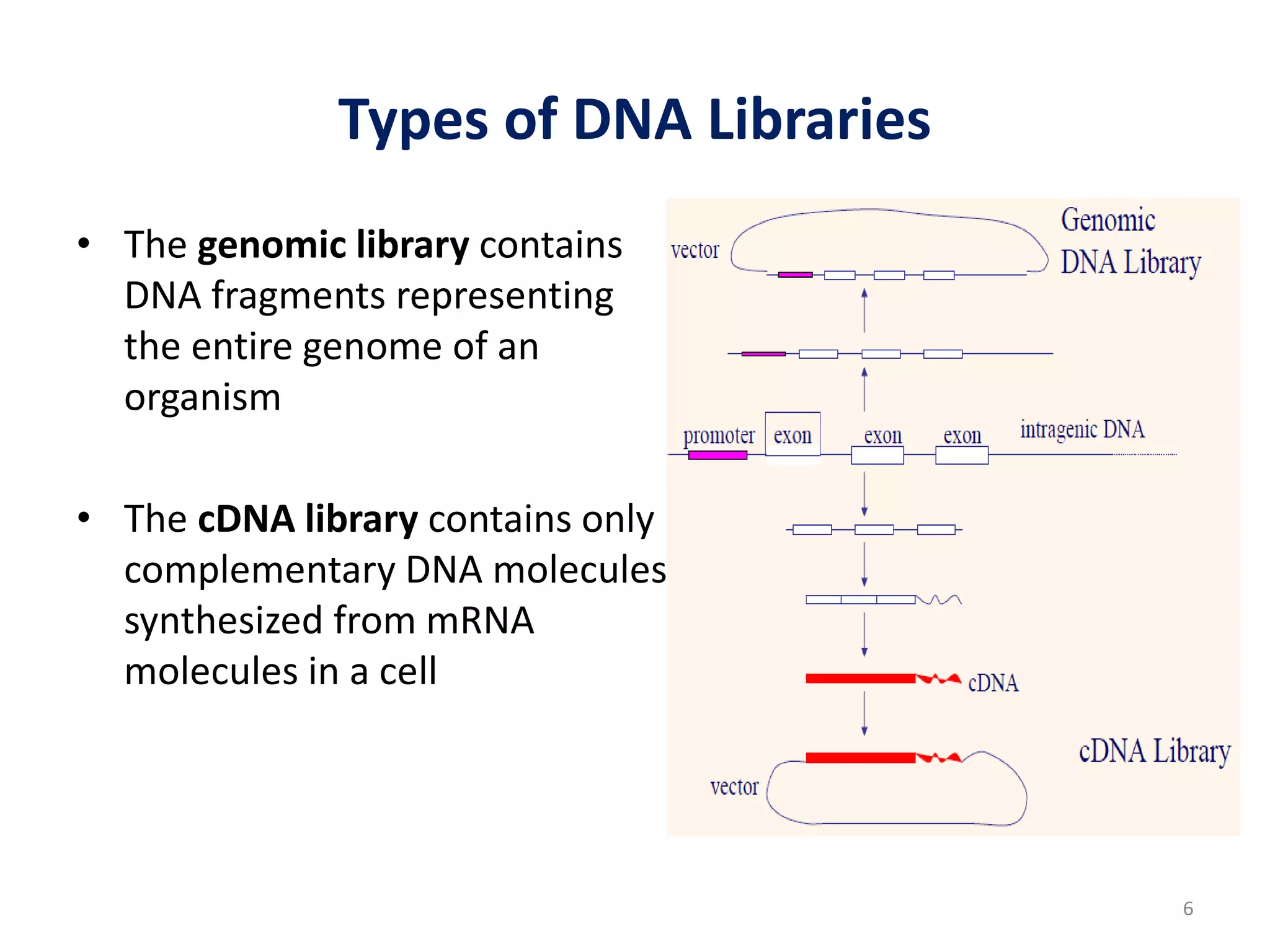
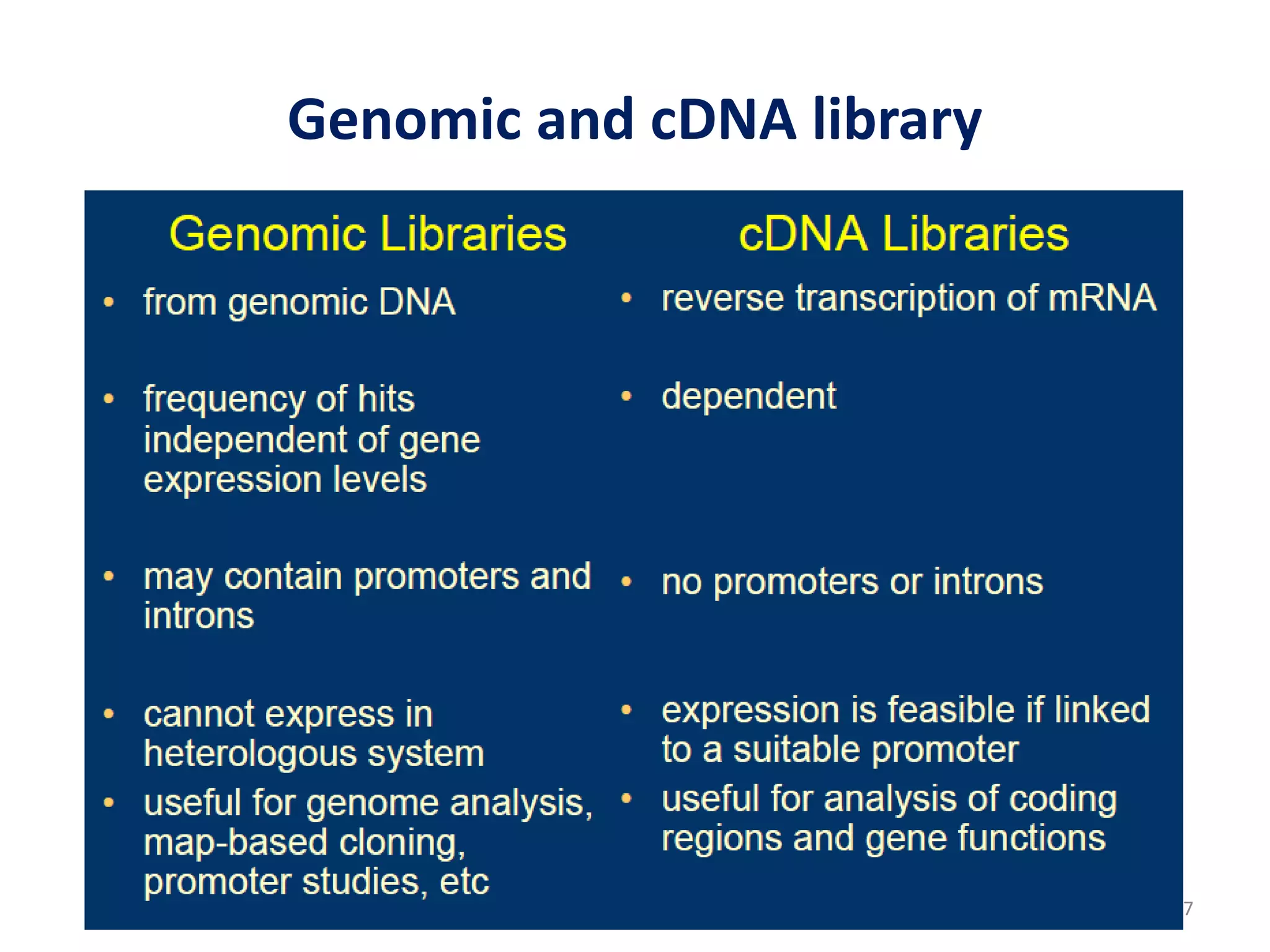
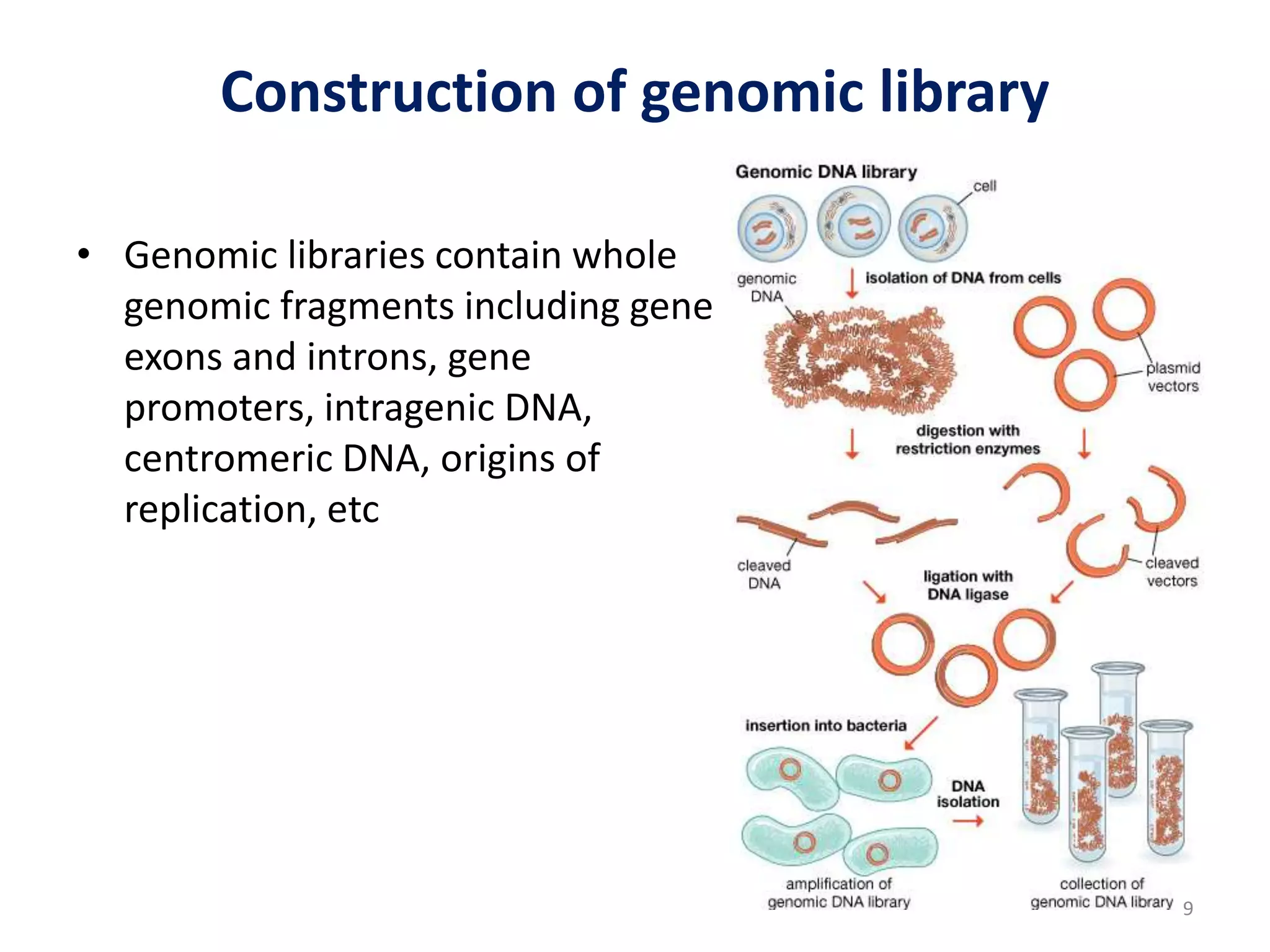
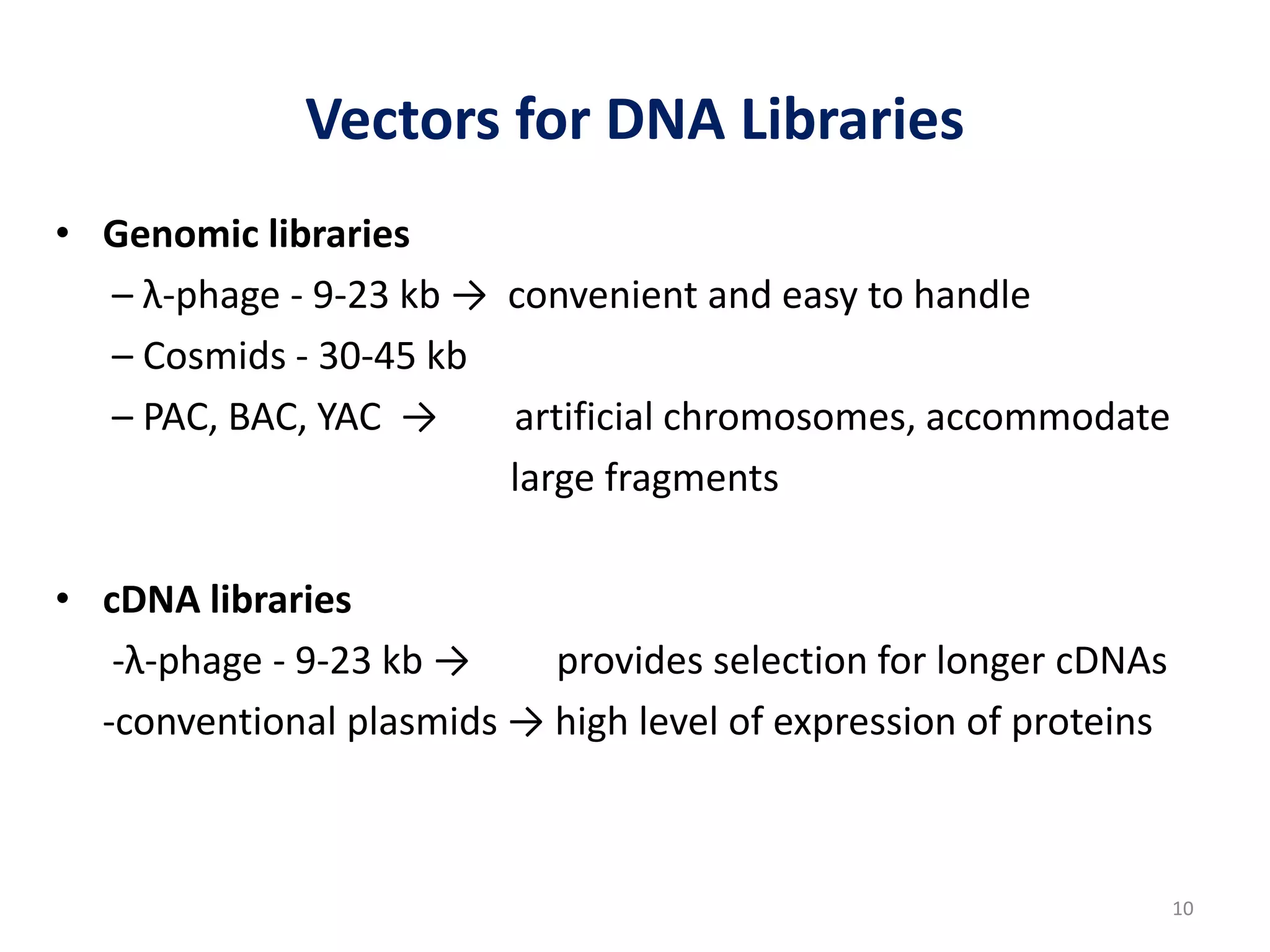
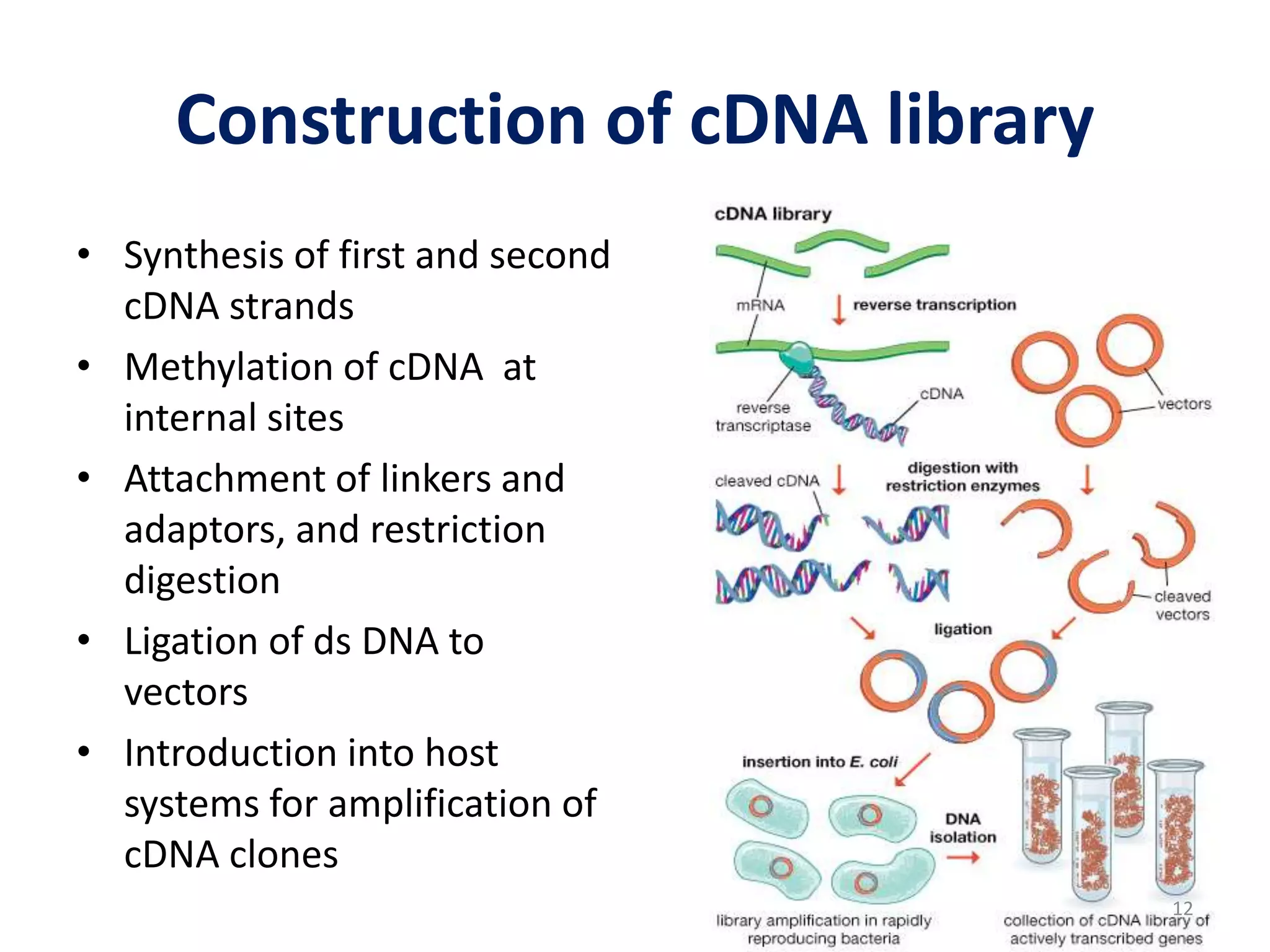
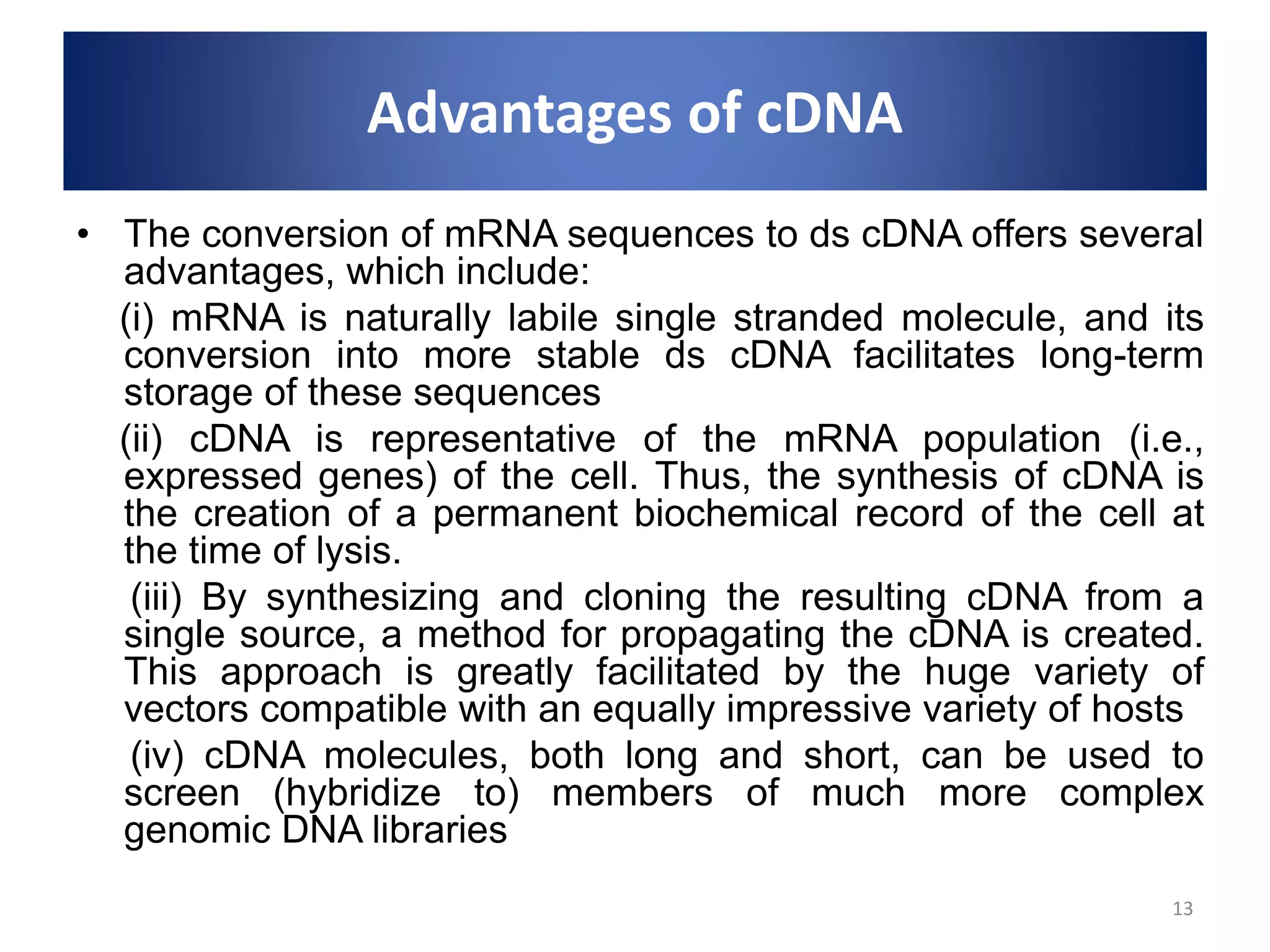
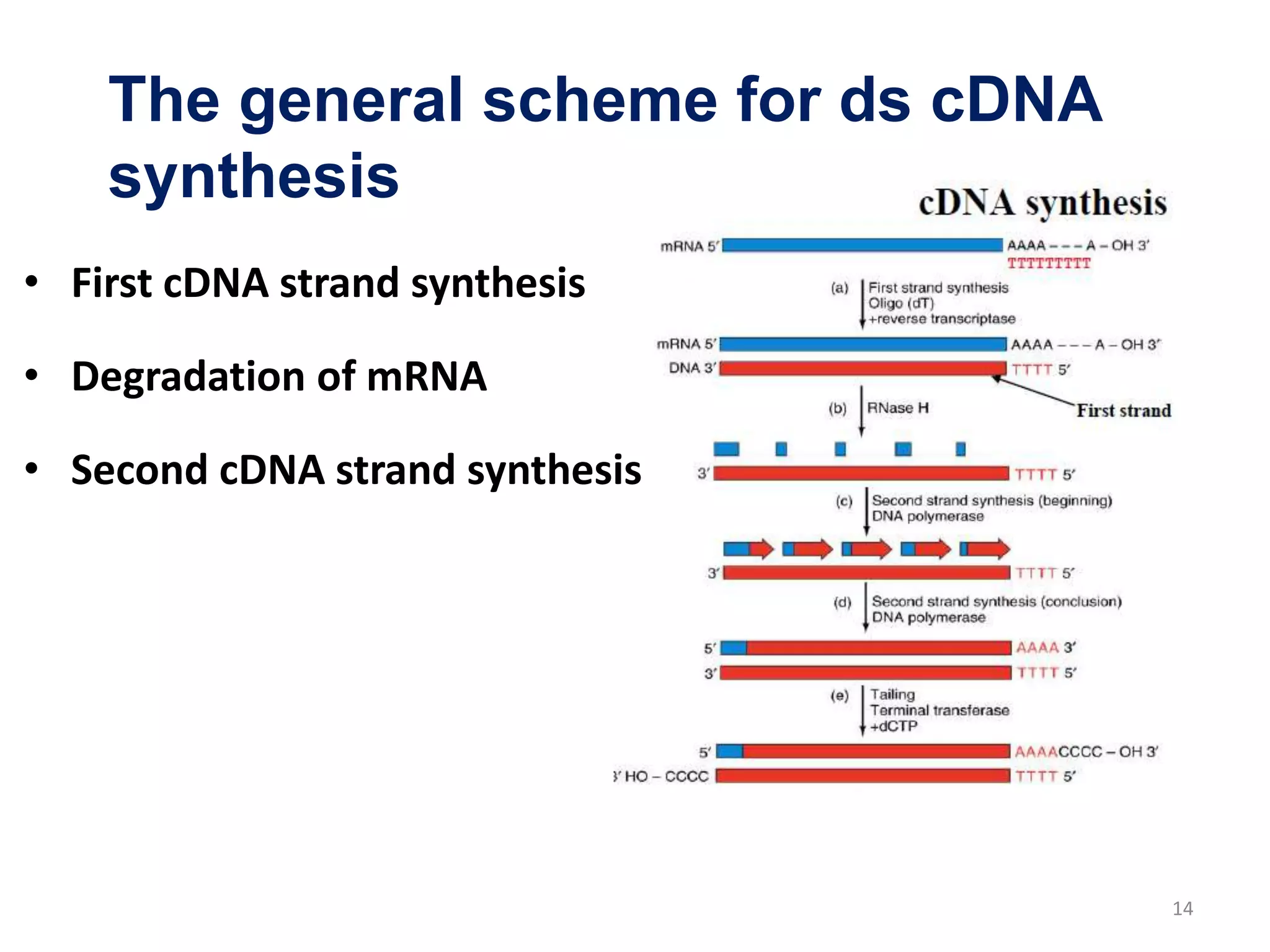
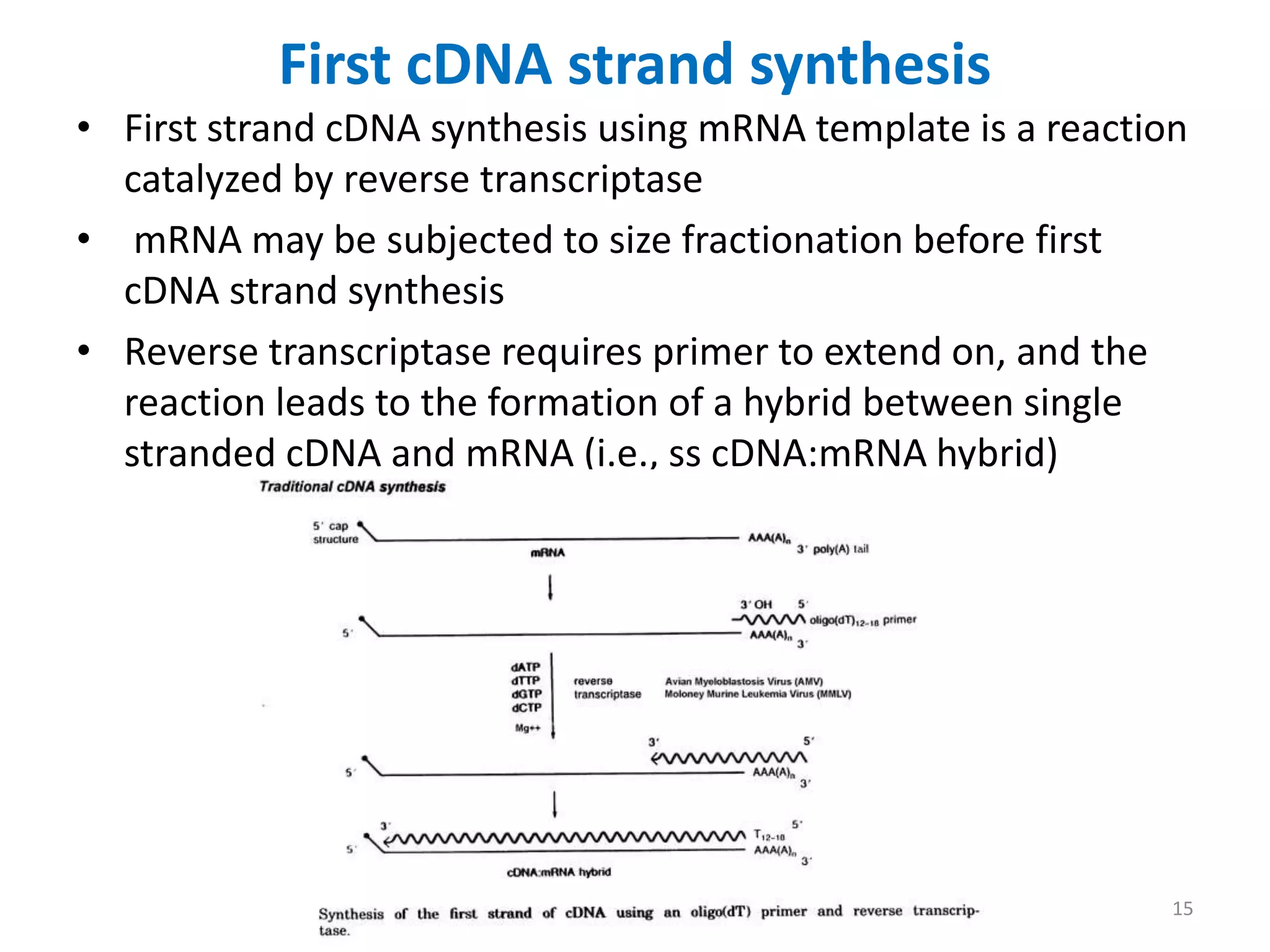
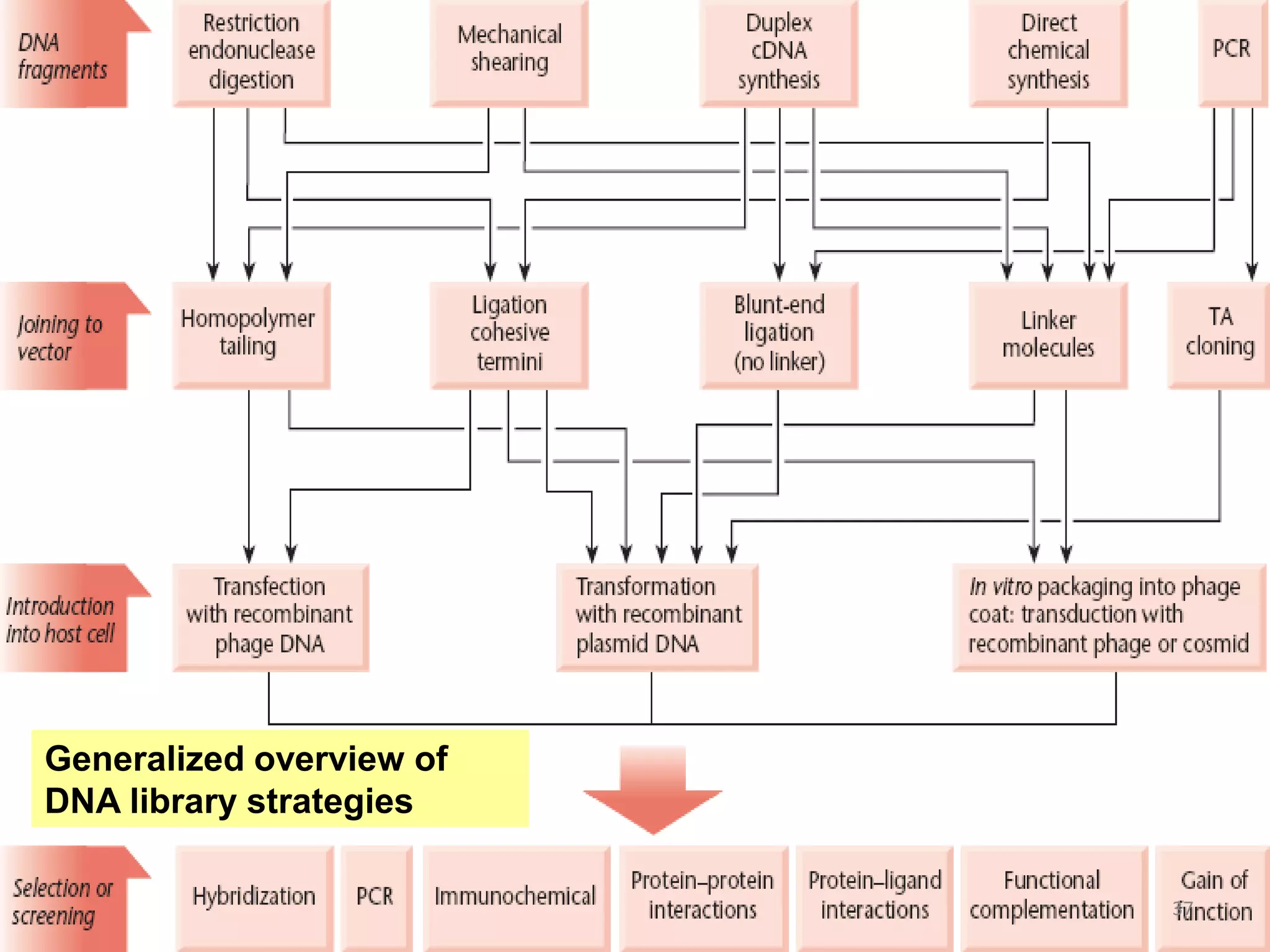
Genomic and cDNA libraries are collections of clones containing DNA fragments from an organism. A genomic DNA library contains all fragments of the genomic DNA, while a cDNA library contains only coding sequences synthesized from expressed mRNA. Genomic libraries are suitable for prokaryotes due to their small genomes but eukaryotic genomes require too many clones, so cDNA libraries are preferred for eukaryotic gene cloning. cDNA libraries represent the expressed genes and contain only coding regions without introns.