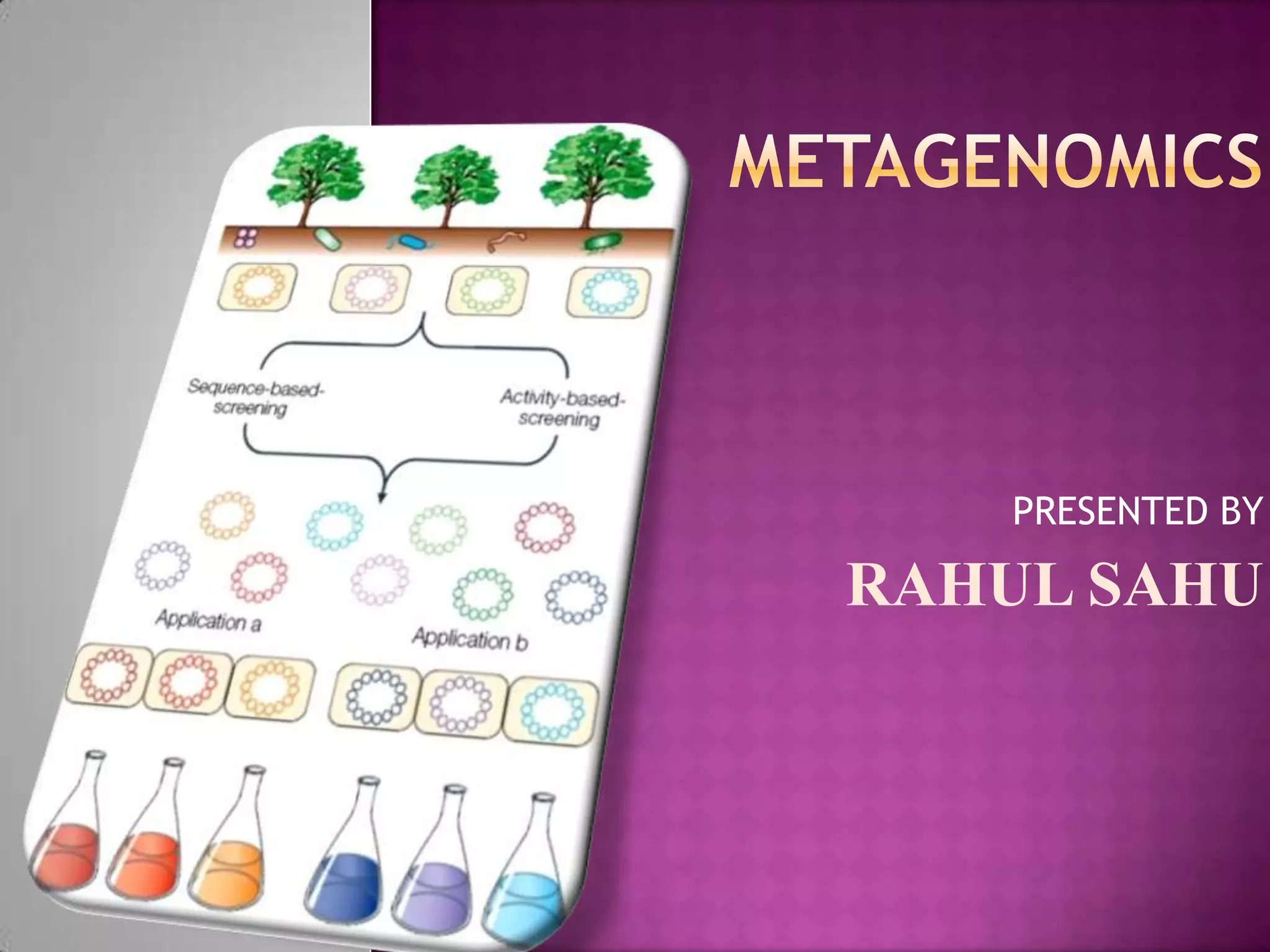
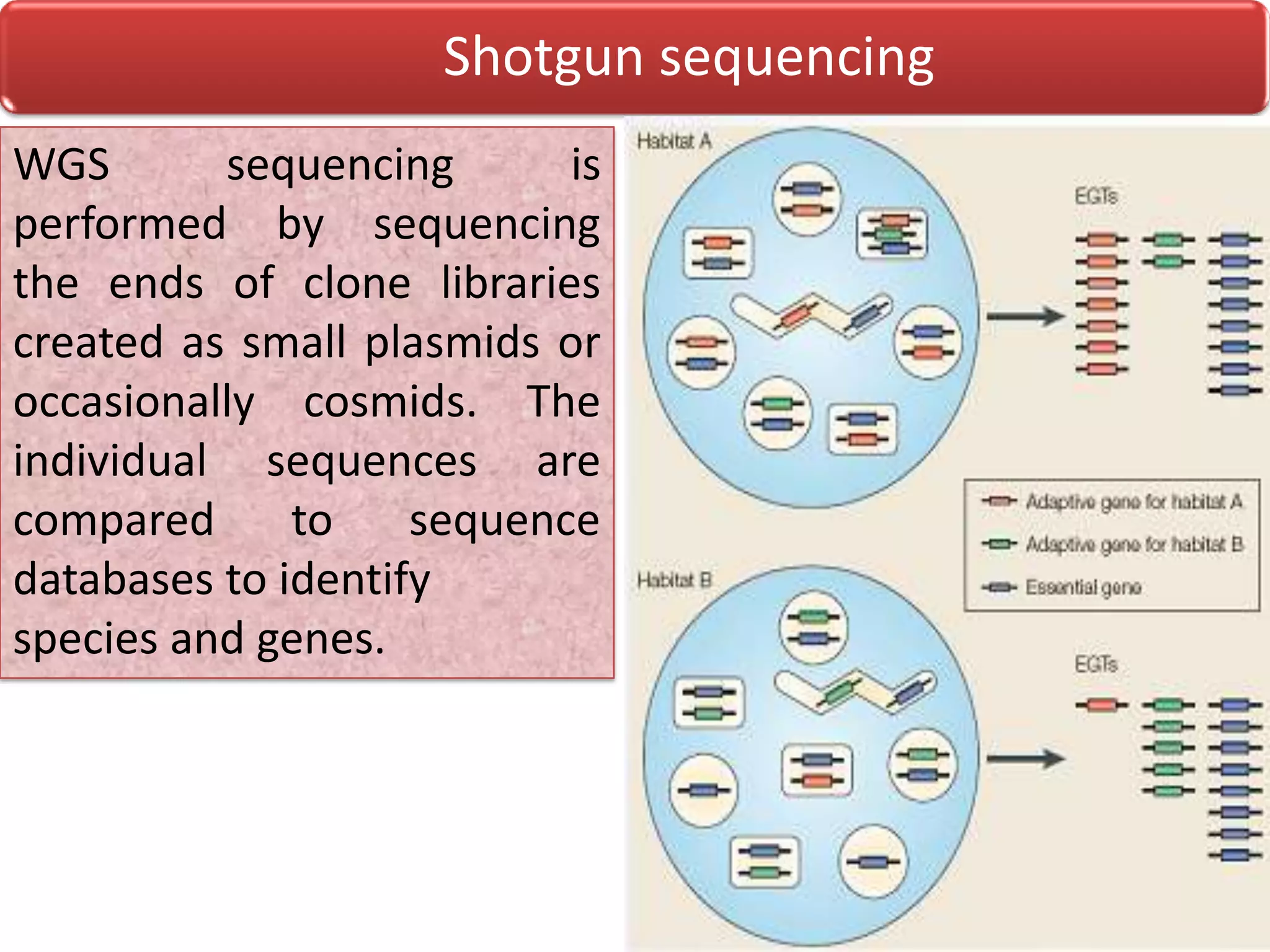
Metagenomics is the study of genetic material recovered directly from environmental samples without culturing. This field enables research on uncultured organisms and microbial communities. There are three main metagenomic approaches: biochemical, whole genome shotgun sequencing, and 16s rRNA sequencing. Metagenomics is being applied to study human microbiomes, discover new genes and enzymes, monitor environmental impacts, and characterize uncultured microbes. Future directions include identifying more novel products from uncultured bacteria and improving culture methods and bioinformatics tools.