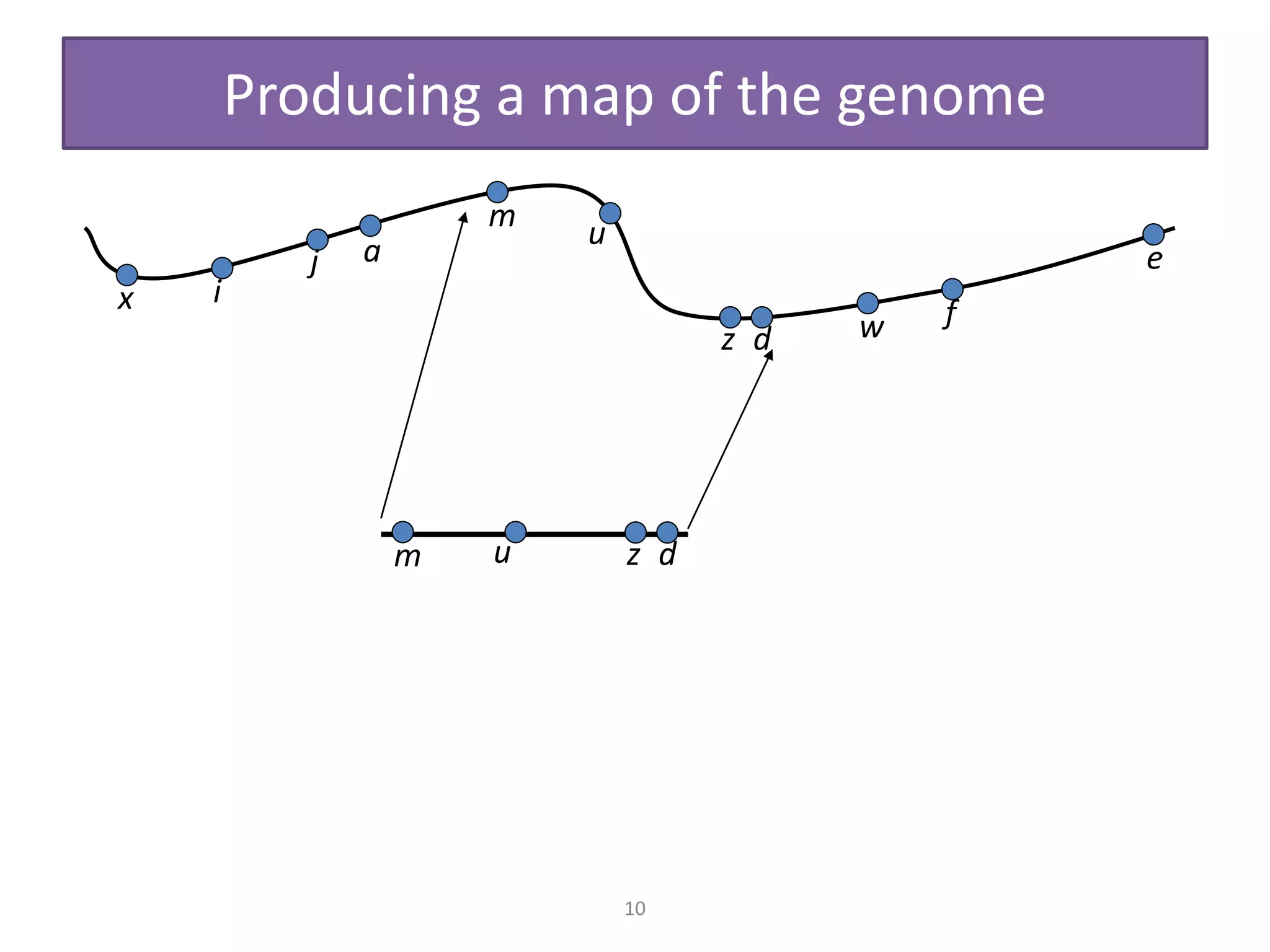
Gene mapping tools can be used to represent the arrangement of genes on chromosomes and determine the position and distance between genes. There are several types of gene mapping including physical mapping, genetic mapping, and linkage mapping. Physical mapping uses molecular techniques to examine DNA directly to construct maps showing gene positions. Genetic maps are based on recombination frequencies between genes and measure distances in centimorgans. Different DNA markers can also be used for mapping, including RFLPs, SNPs, SSLPs, and STSs, along with techniques like FISH, restriction digests, and PCR to analyze these markers and produce genome maps. Gene mapping is important for identifying mutant genes that cause genetic disorders and furthering our understanding of human genetics.