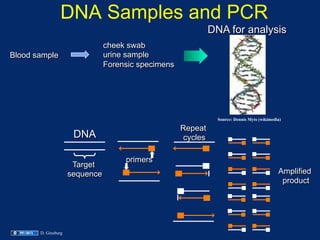
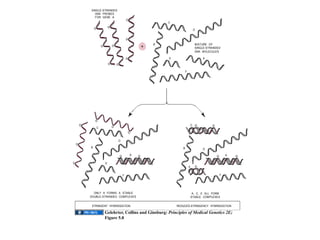
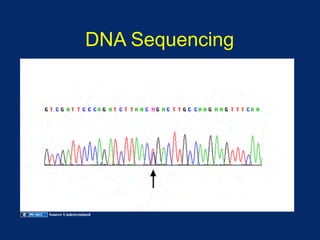
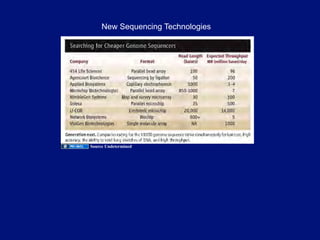
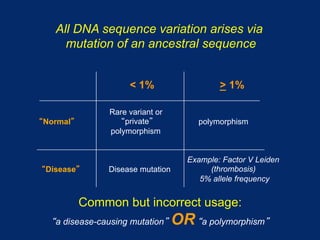
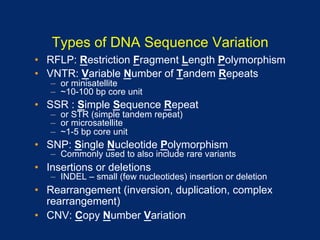
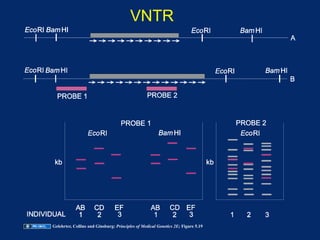
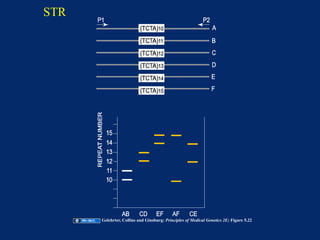
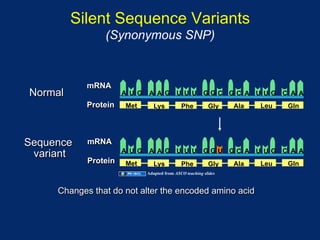
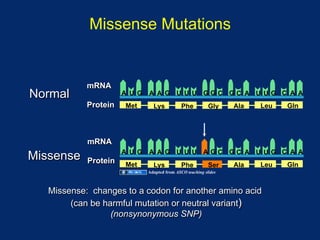
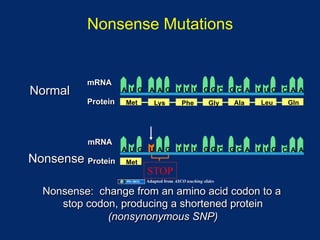
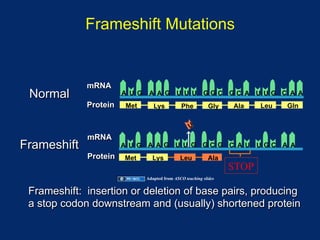
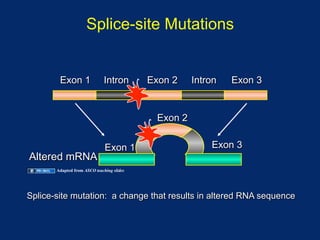
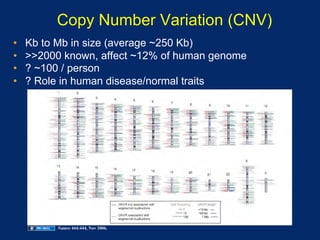
The document discusses DNA sequence variation, focusing on different types of polymorphisms and mutations, including their definitions, classifications, and implications for diseases. It outlines the importance of distinguishing between disease-causing mutations and neutral variations, and emphasizes understanding polymorphisms such as SNPs and CNVs. The text also provides insight into the methodologies used for detecting mutations and the relevance of genetic variations in the context of human health.