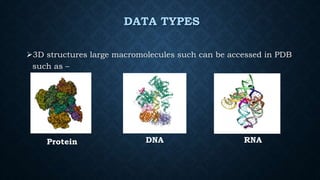
The Protein Data Bank (PDB) is an online archive of experimentally determined 3D structures of biological macromolecules, primarily proteins and nucleic acids, accessible to researchers and the public. Established in 1971 and now managed by the Research Collaboratory for Structural Bioinformatics, PDB utilizes various structure determination techniques and formats for data exchange. Its applications span drug discovery, structural genomics, and education, making it an essential resource in structural biology.