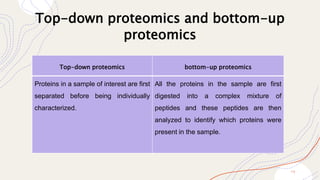
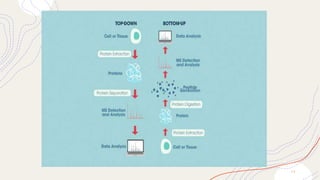
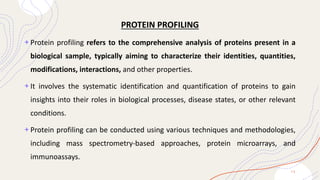
Proteomics is the comprehensive study of proteomes, which are the entire set of proteins expressed by an organism under specific conditions, and can vary between different cell types and stages. This field includes various types of proteomic analyses such as structural, functional, and expression proteomics, each focusing on different aspects of protein characteristics and dynamics. Techniques like mass spectrometry and protein profiling are essential for identifying, quantifying, and understanding protein interactions and modifications, playing a crucial role in advancing biomedical research, including cancer studies.