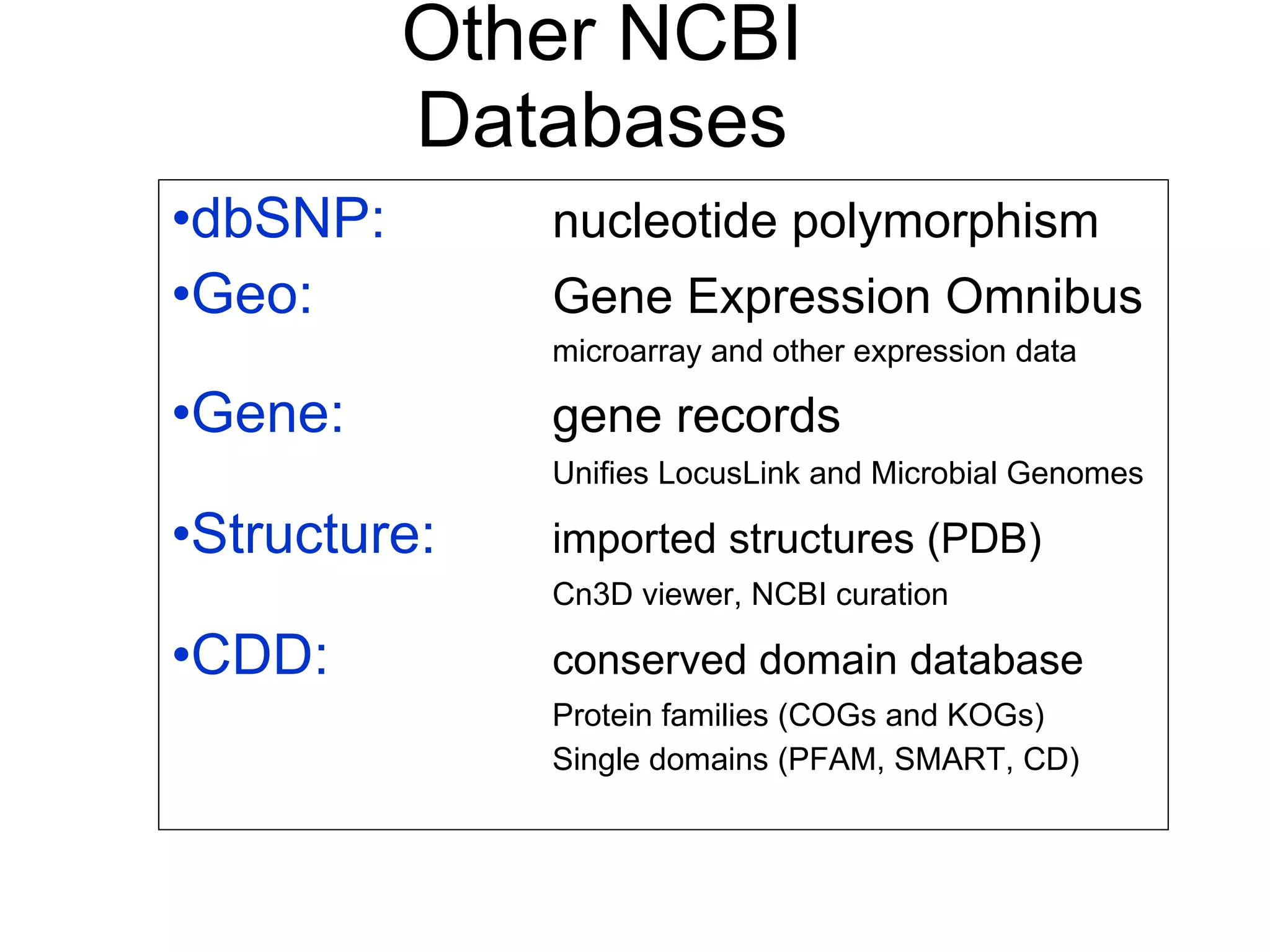
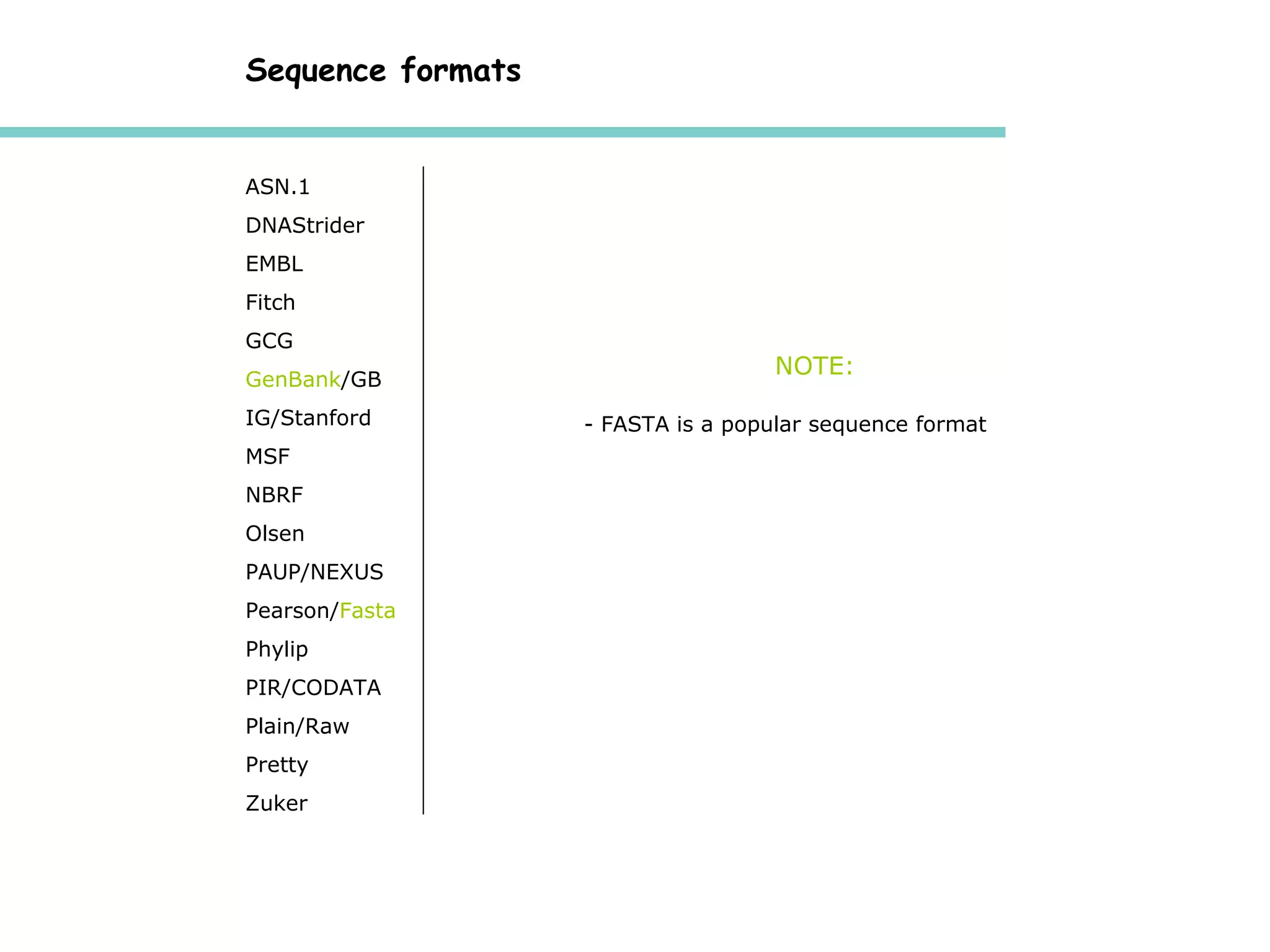
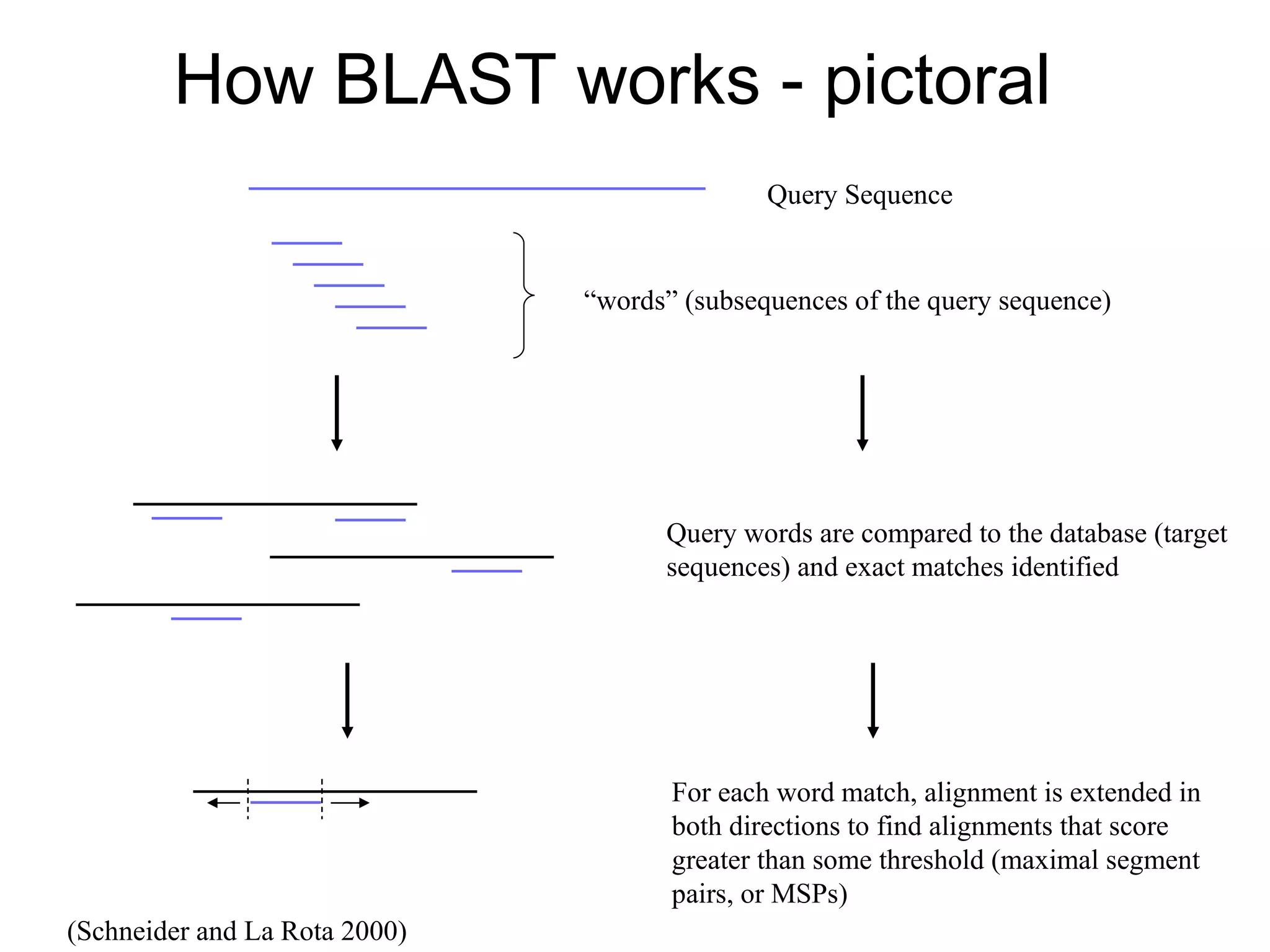
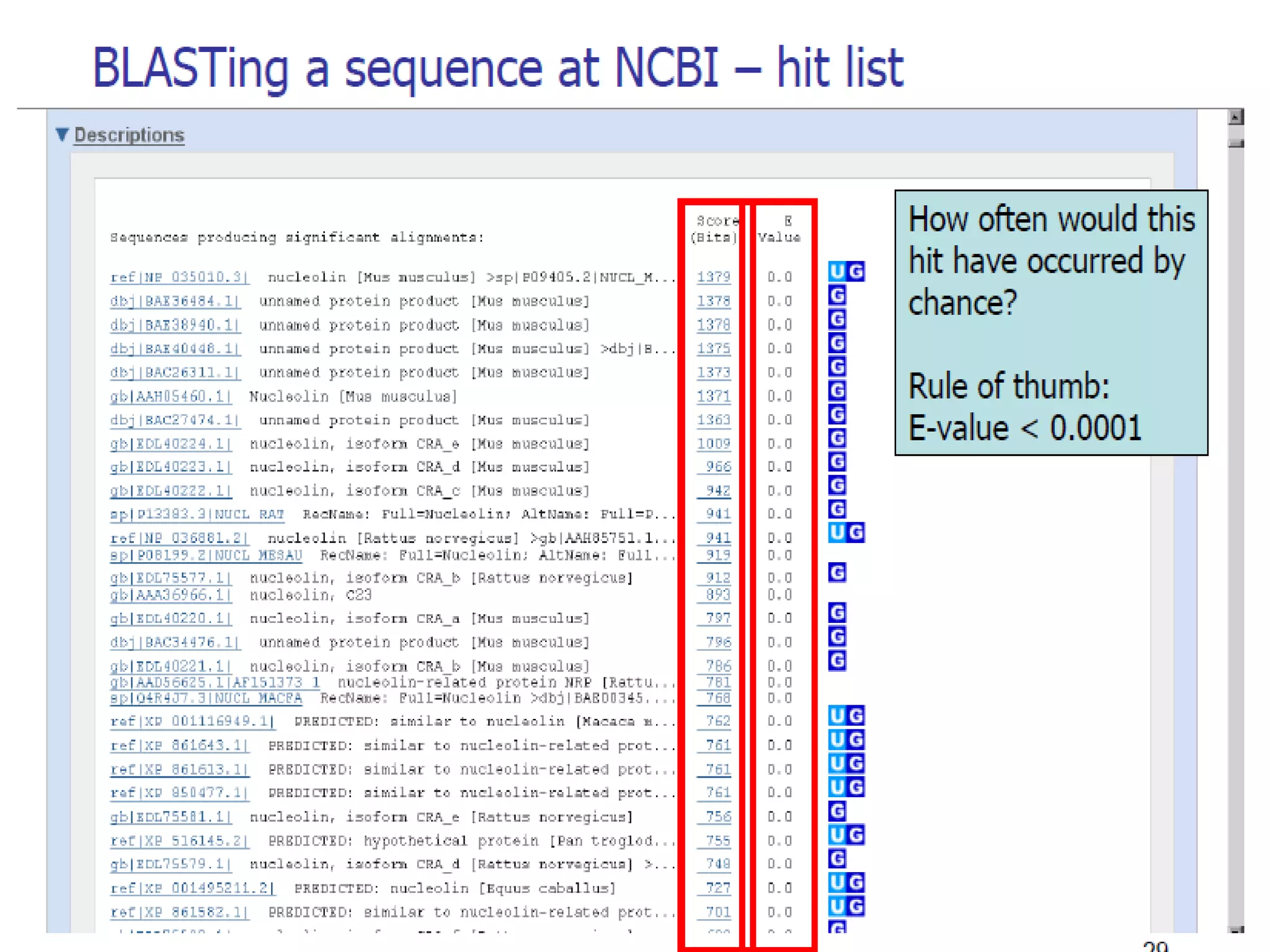
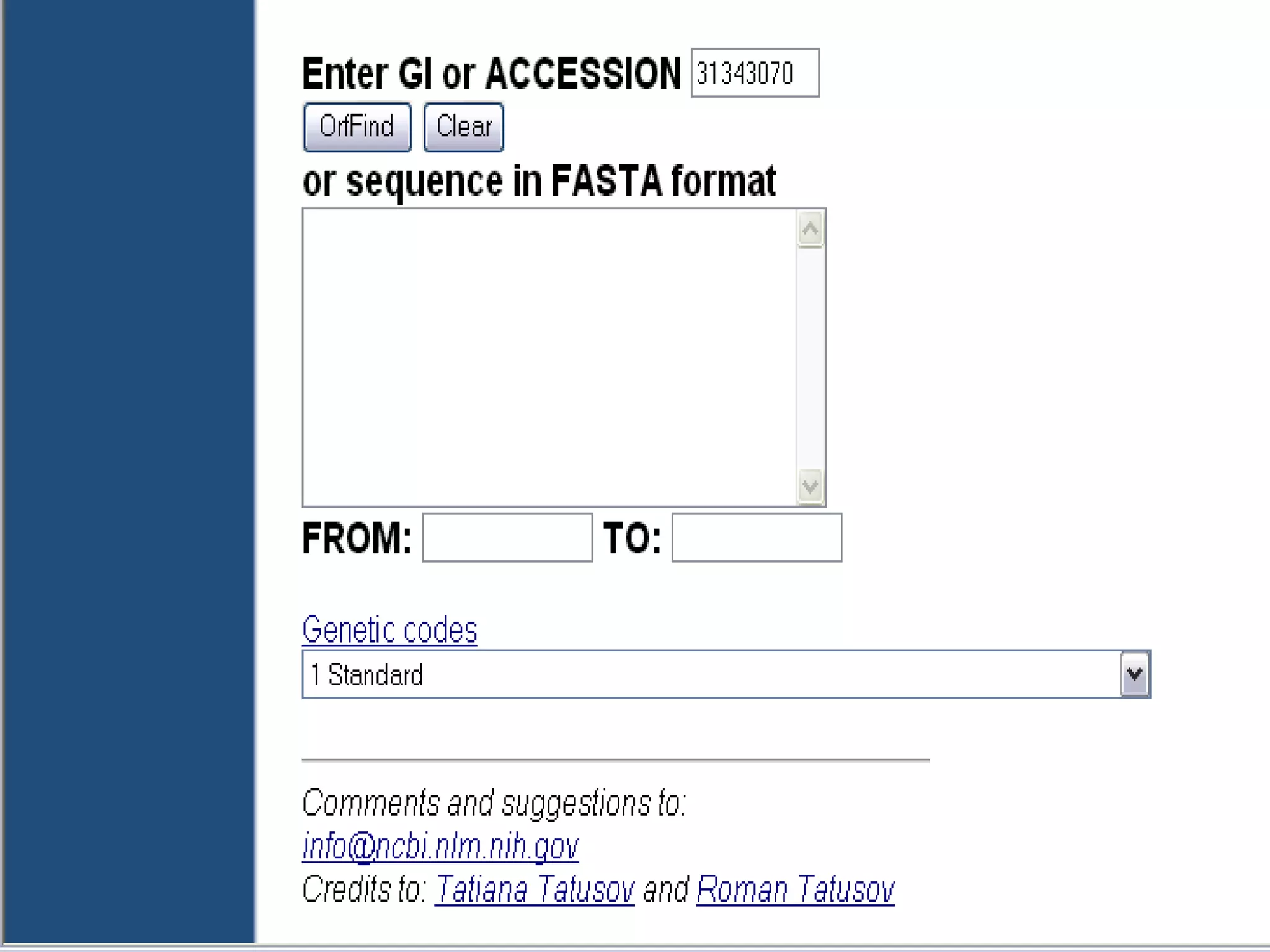
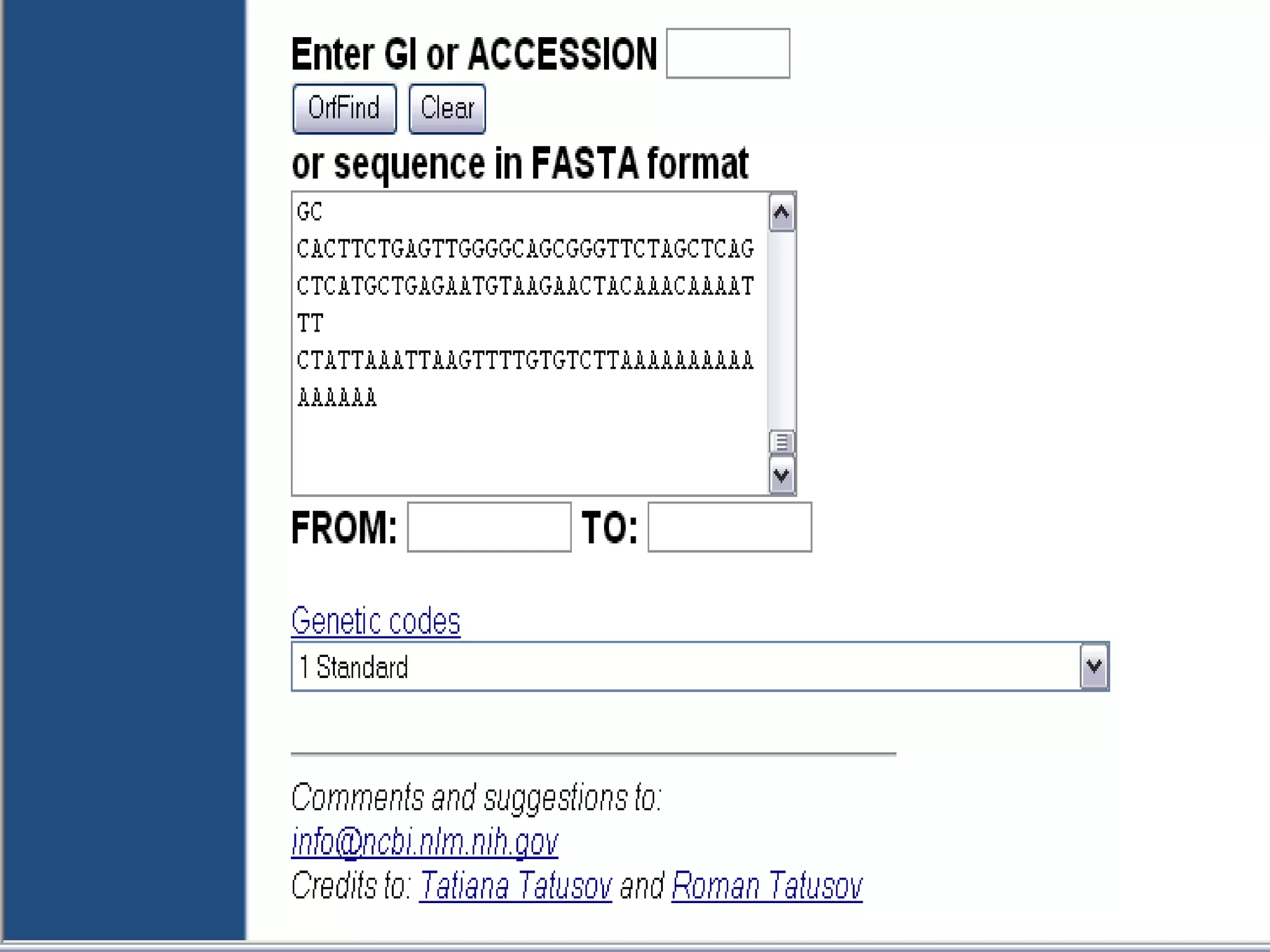
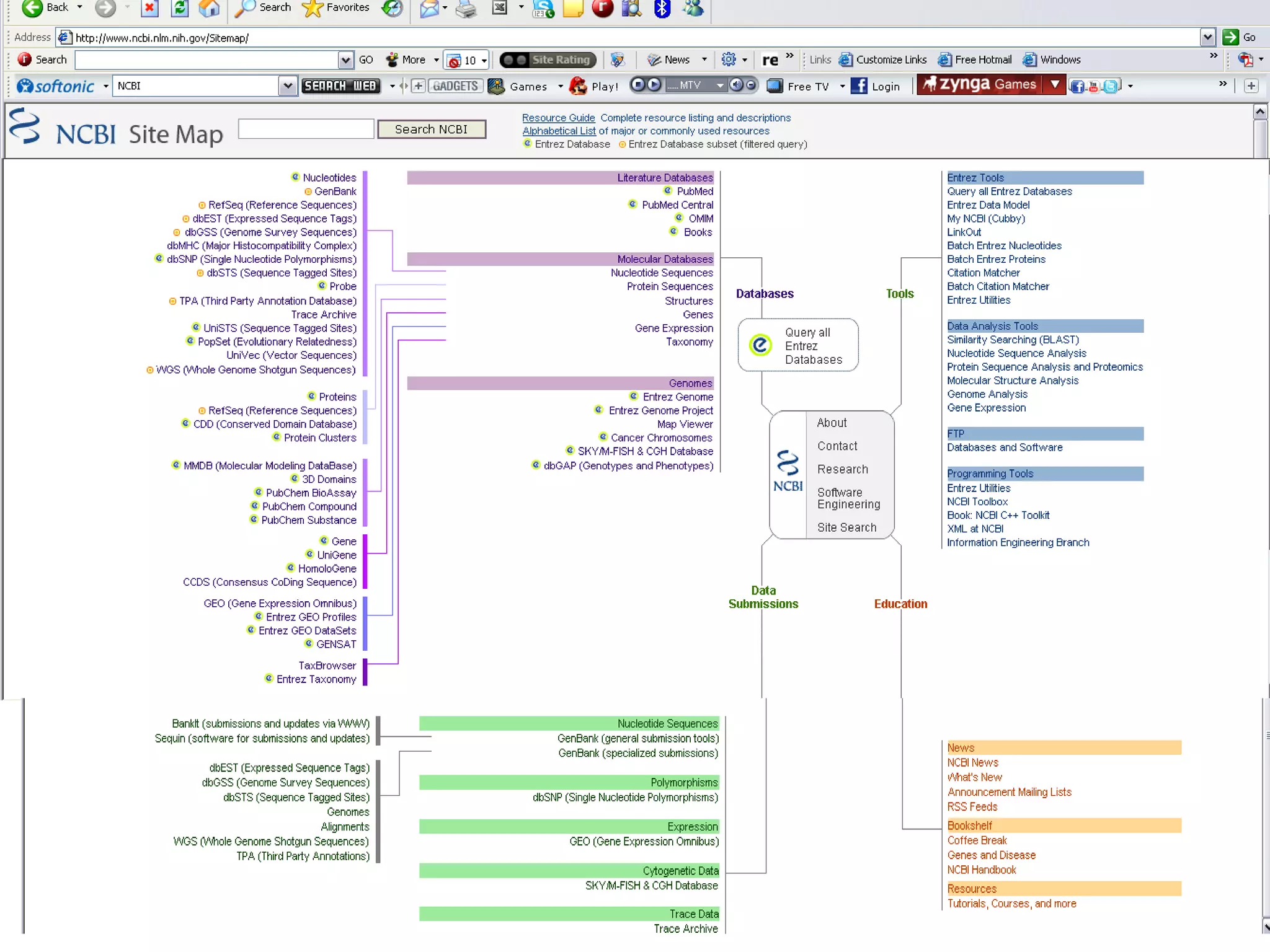
The National Center for Biotechnology Information (NCBI) was created in 1988 as part of the National Library of Medicine at NIH. It establishes public databases for biological research, develops software tools for sequence analysis, and disseminates biomedical information from its location in Bethesda, MD. NCBI houses several integrated databases including PubMed, GenBank, RefSeq, and UniGene that contain literature, sequences, gene information, and more.







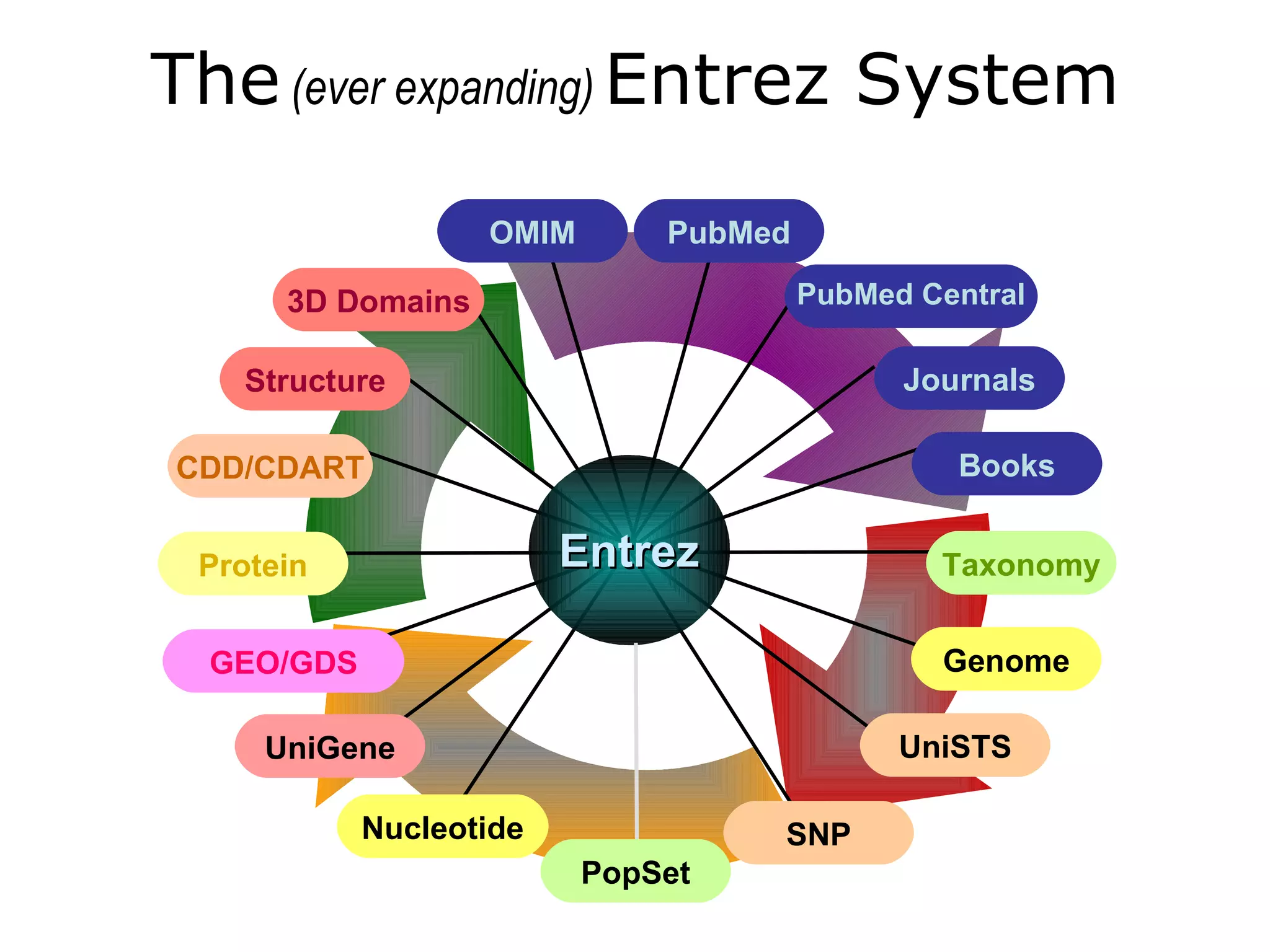
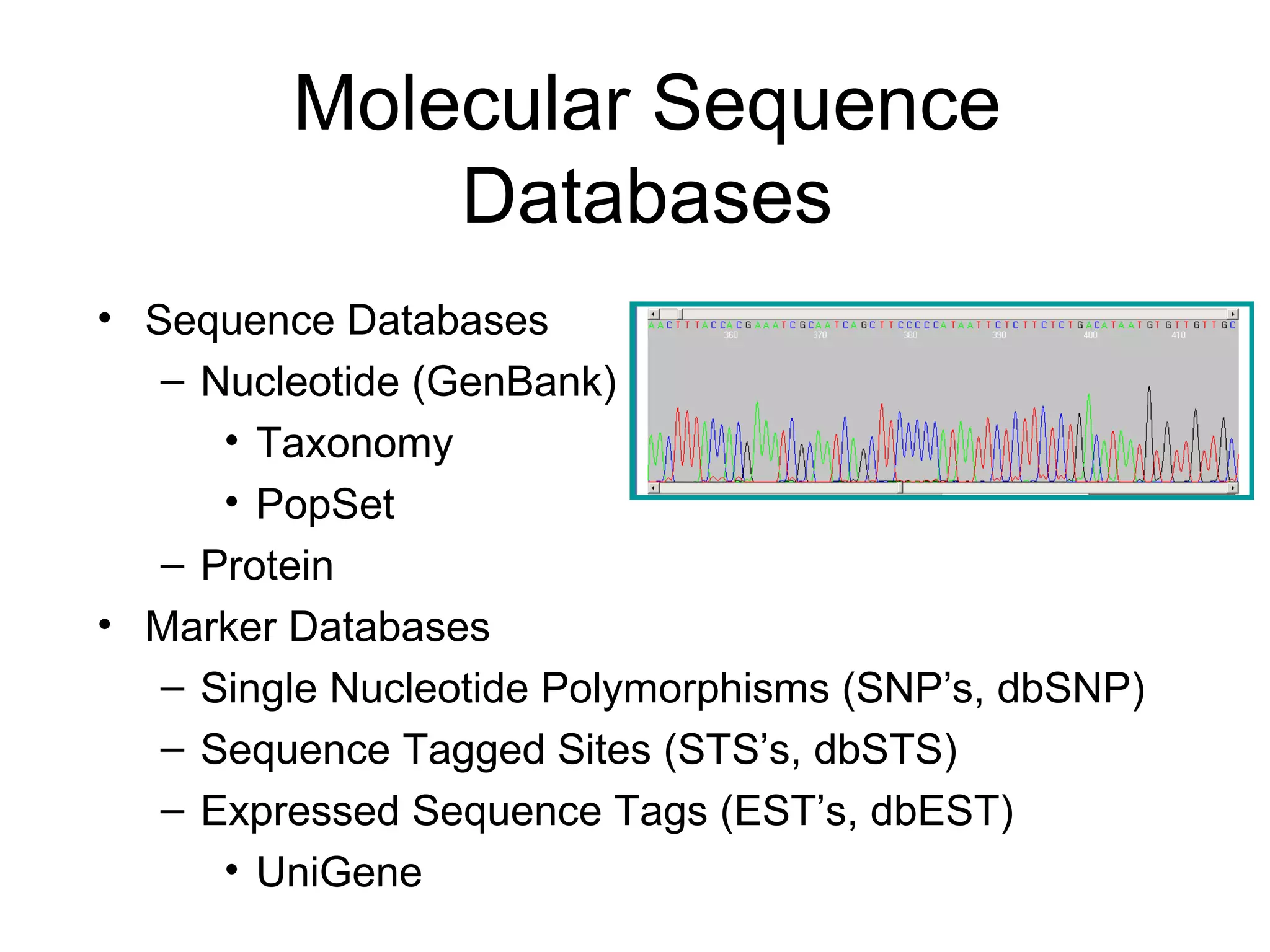
![Entrez Databases All Molecular Database entries are organized by organism (Taxonomy Database) . Each record is assigned a UID. A “unique integer identifier” for internal tracking Each record is indexed by data fields. [author], [title], [organism], and many others Each record is given a Document Summary. a summary of the record’s content (DocSum) Each record is manually or computationally assigned links to biologically related UIDs in and across databases.](https://image.slidesharecdn.com/ncbifinal-111218021720-phpapp01/75/NCBI-10-2048.jpg)











![Whole Genome Shotgun (WGS) Projects wgs master[properties] ftp://ftp.ncbi.nih.gov/genbank/wgs/](https://image.slidesharecdn.com/ncbifinal-111218021720-phpapp01/75/NCBI-24-2048.jpg)




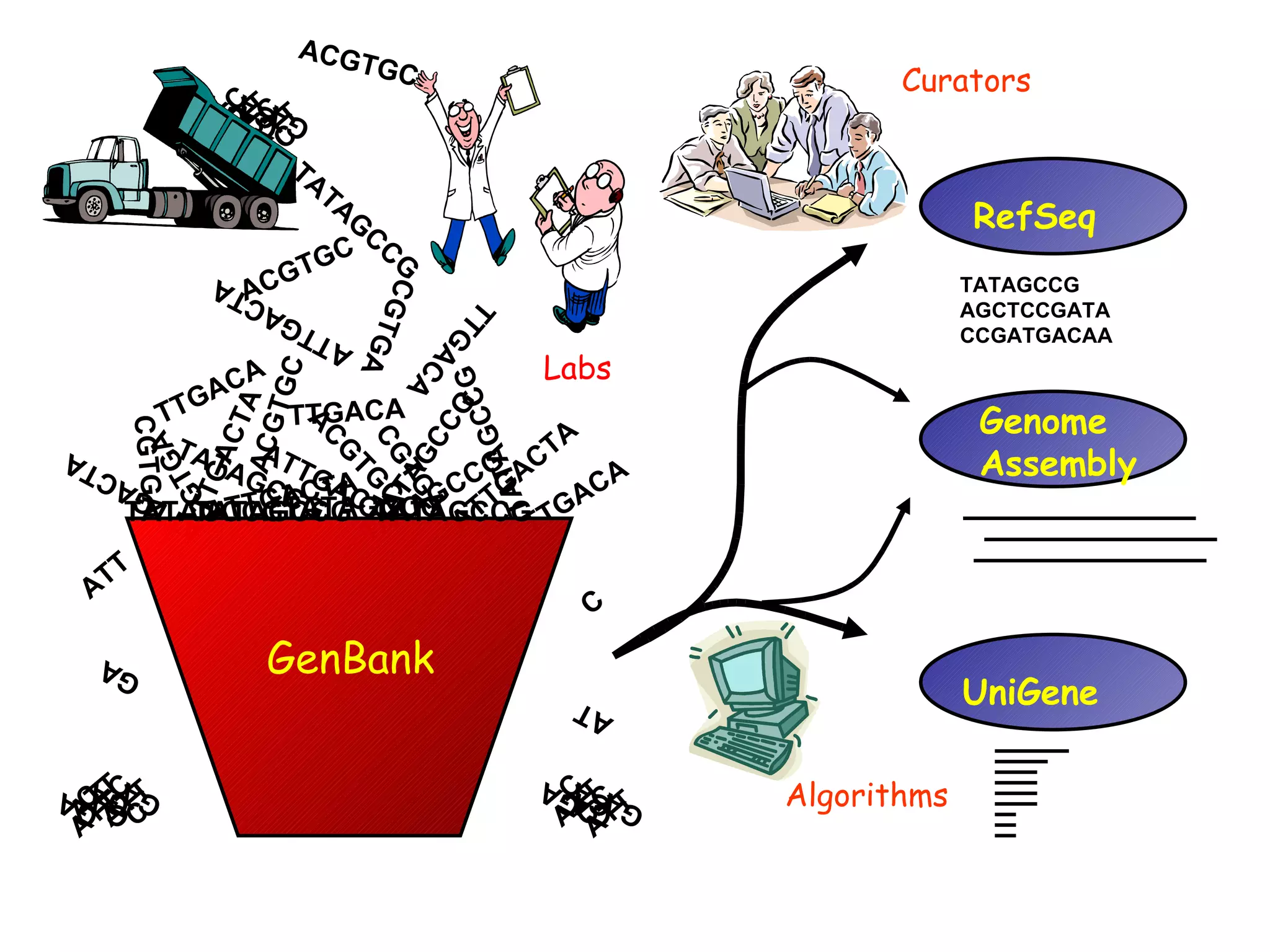
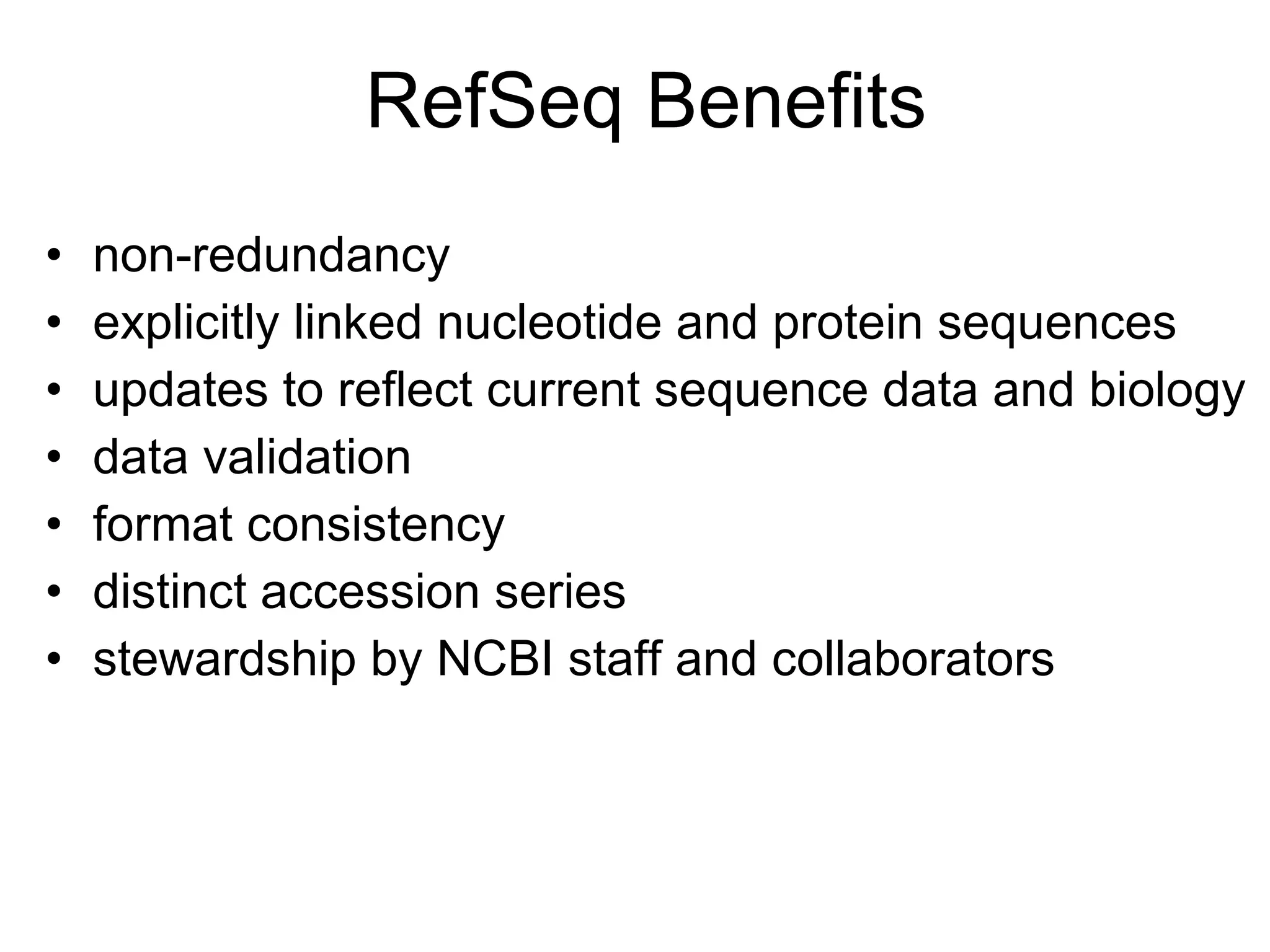
![RefSeq: NCBI’s Derivative Sequence Database Curated transcripts and proteins reviewed human, mouse, rat, fruit fly, zebrafish, arabidopsis Model transcripts and proteins Assembled Genomic Regions (contigs) human genome mouse genome Chromosome records Human genome microbial organelle ftp://ftp.ncbi.nih.gov/refseq/release / srcdb_refseq[Properties]](https://image.slidesharecdn.com/ncbifinal-111218021720-phpapp01/75/NCBI-30-2048.jpg)

![Third Party Annotation (TPA) Database Annotations of existing GenBank sequences Allows for community annotation of genomes Direct submissions BankIt Sequin tpa[Properties]](https://image.slidesharecdn.com/ncbifinal-111218021720-phpapp01/75/NCBI-33-2048.jpg)