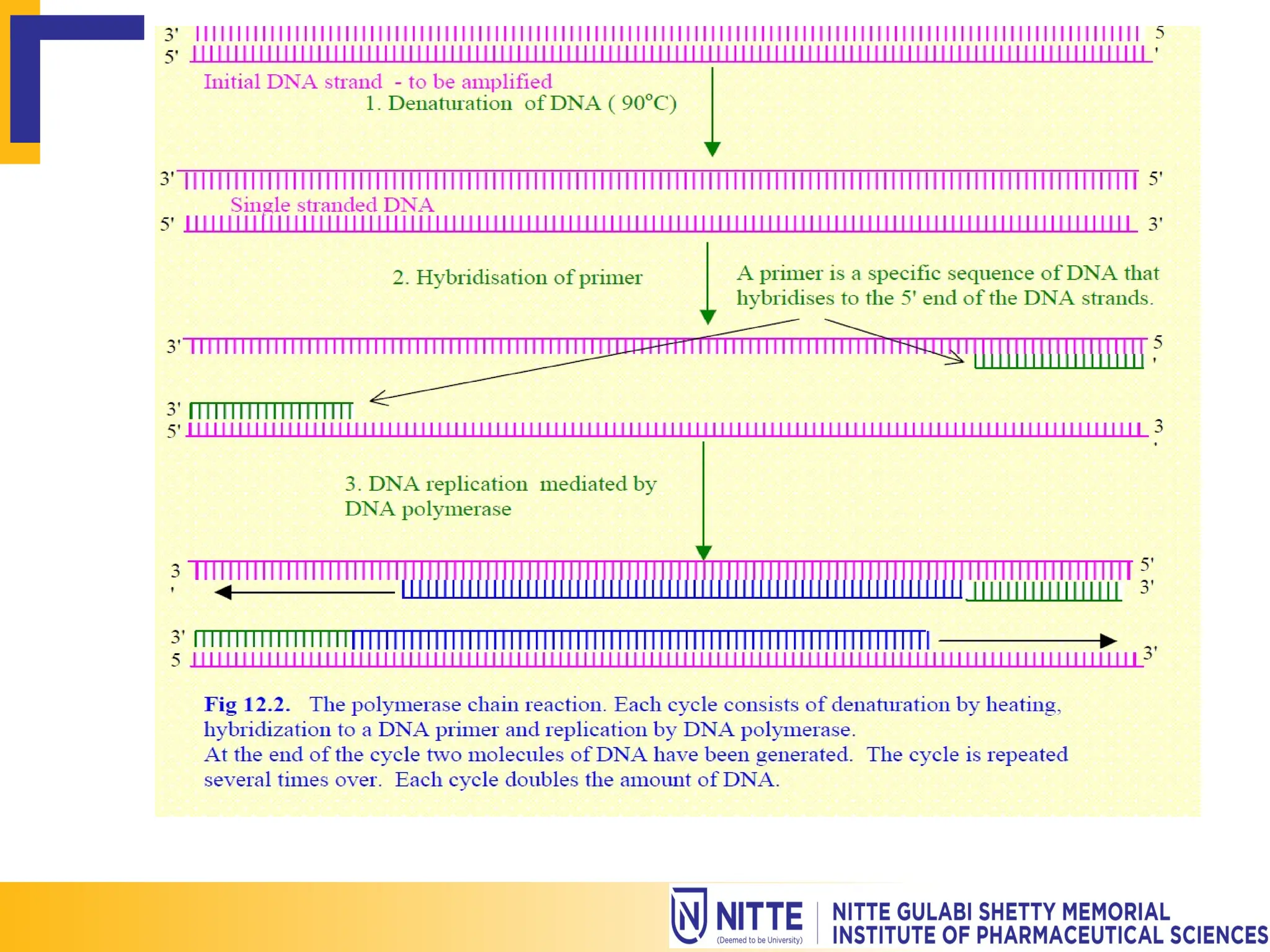
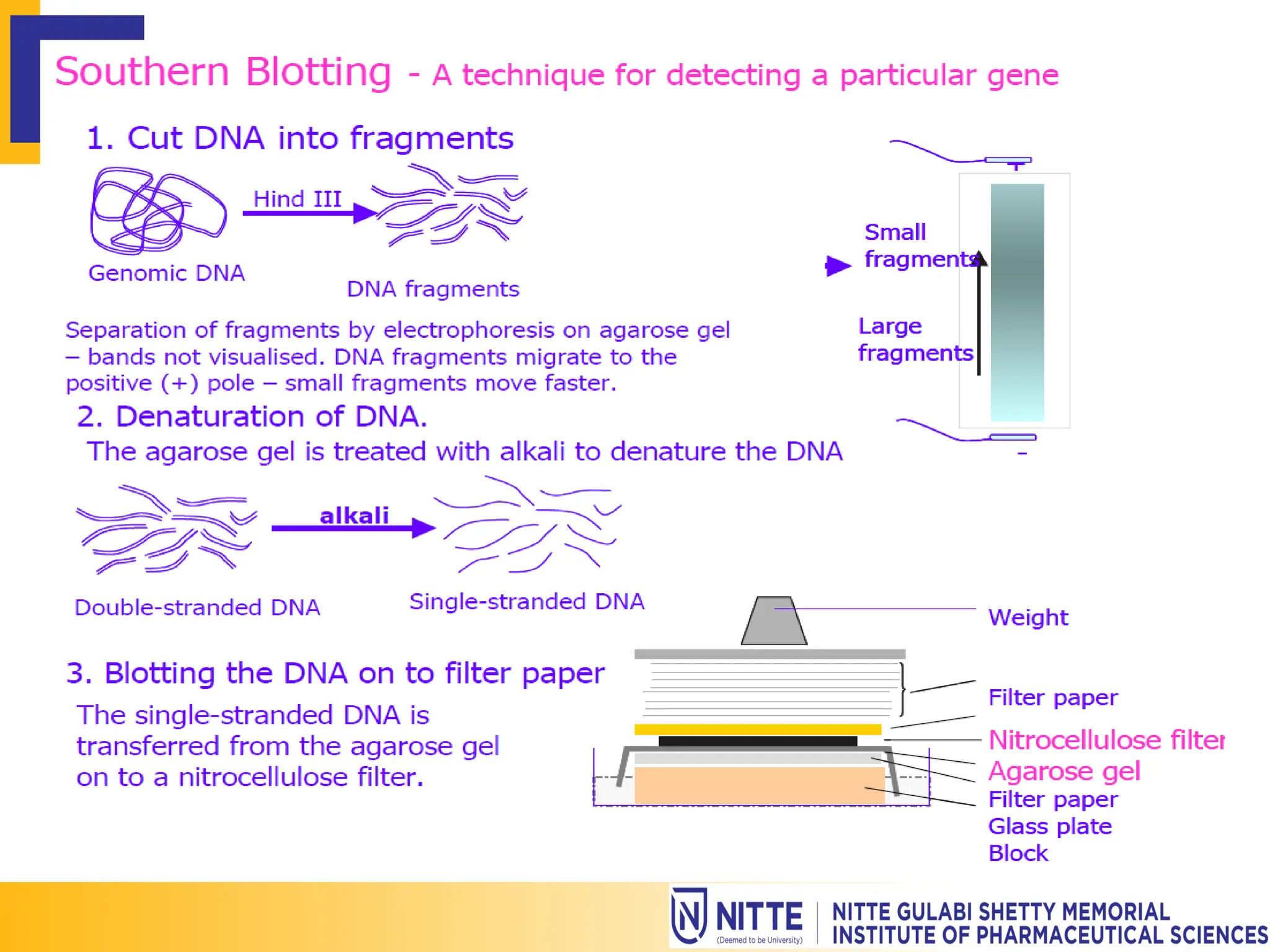
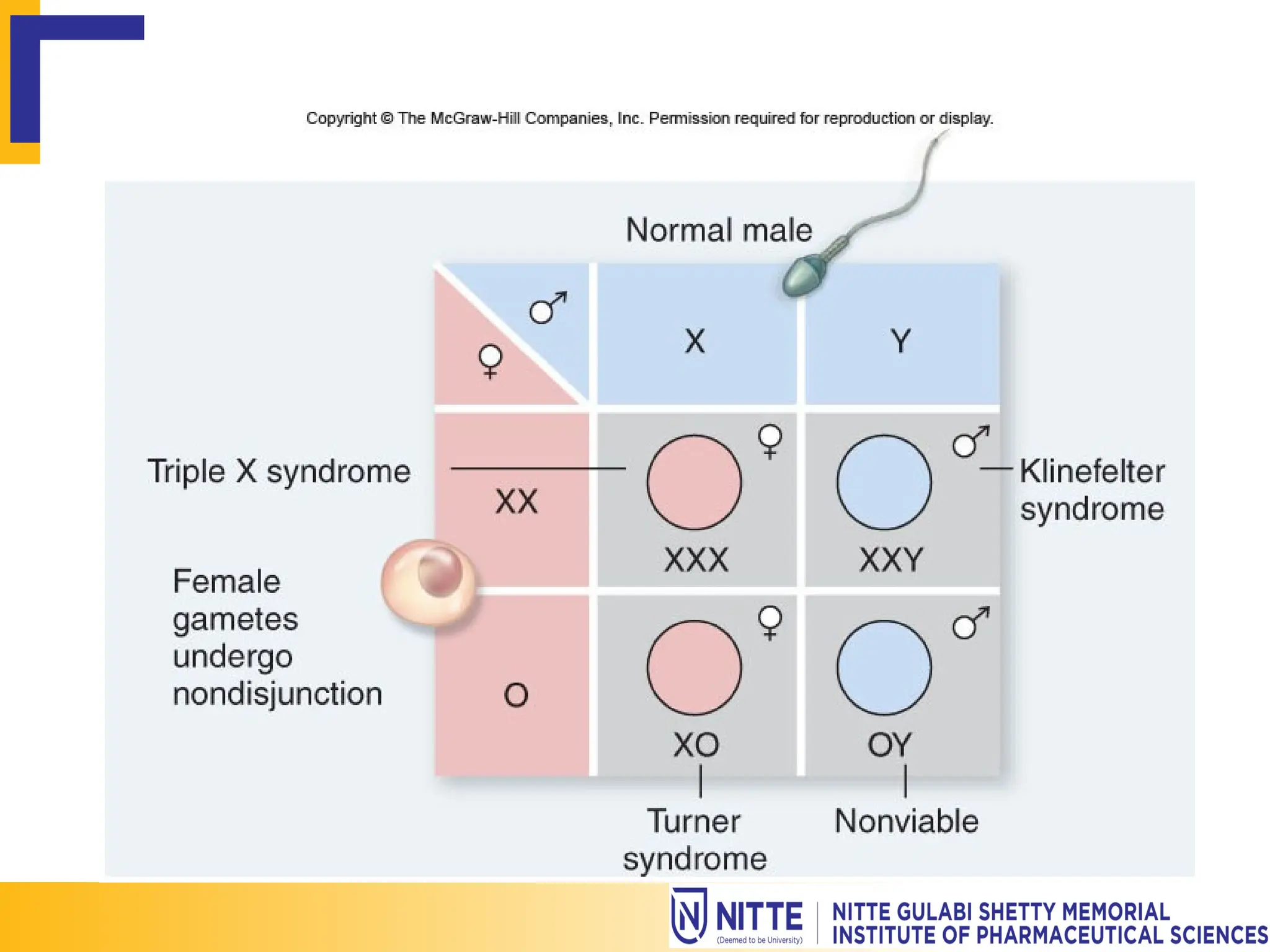
Gene mapping, also known as genome or linkage mapping, is the process of creating a genetic map that links DNA fragments to their respective chromosomes, crucial for understanding genetic diseases. This document delves into various gene mapping techniques, including genetic, physical, and cytogenetic mapping, as well as tools like RFLP and PCR for identifying gene locations. Additionally, it highlights how gene mapping aids in identifying inherited disorders caused by altered proteins or abnormal chromosome numbers.