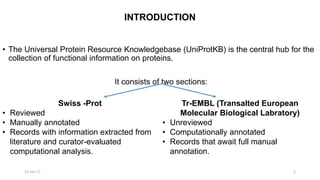
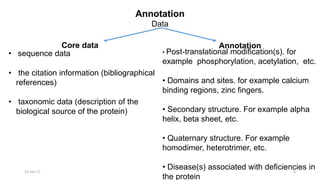
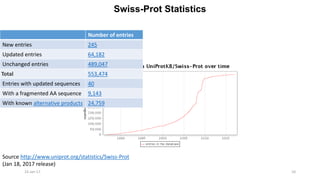
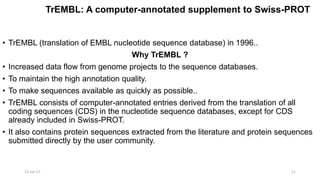
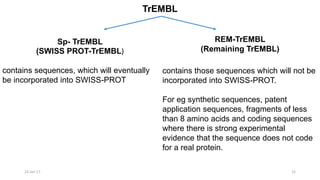
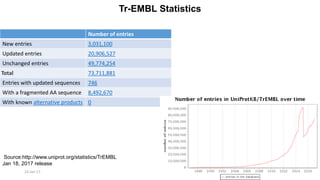
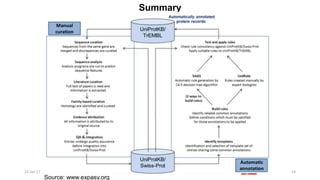
The document provides an overview of the Universal Protein Resource Knowledgebase (UniProtKB), detailing its two main components, Swiss-Prot and TrEMBL, which contain manually annotated and computationally annotated protein information, respectively. It emphasizes the high level of annotation, minimal redundancy, and integration with other databases, while also presenting statistics regarding the number of entries. Additionally, it discusses the continuous enhancement of Swiss-Prot's format and content to support proteomics and the use of automated procedures while maintaining annotation quality.