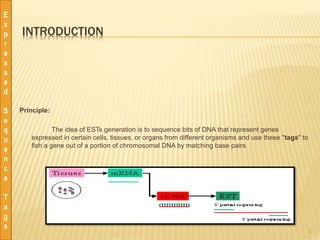
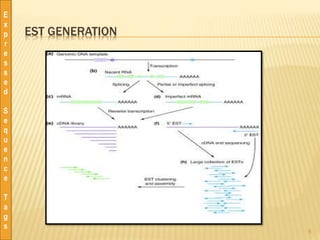
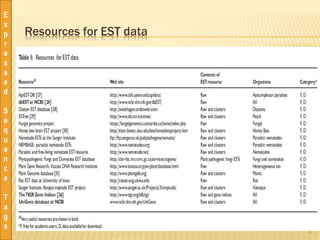
This document discusses expressed sequence tags (ESTs), which are short sequences of cDNA used to identify genes and study gene expression. It provides a brief history of ESTs, noting they were first coined in 1991. ESTs are generated by sequencing fragments of cDNA from mRNA. They provide a quick and inexpensive way to discover new genes and study transcriptomes. Large databases of ESTs exist that can be searched and mined for various applications, including gene discovery, similarity searching, and transcriptome analysis. Pre-processing and clustering/assembling tools are used to improve EST data quality.
![MOLECULAR MARKER
EXPRESSED SEQUENCE TAGS [EST]
By
KAUSHAL KUMAR SAHU
Assistant Professor (Ad Hoc)
Department of Biotechnology
Govt. Digvijay Autonomous P. G. College
Raj-Nandgaon ( C. G. )](https://image.slidesharecdn.com/expressedsequencetagsest-200510110634/75/Expressed-sequence-tag-EST-molecular-marker-1-2048.jpg)