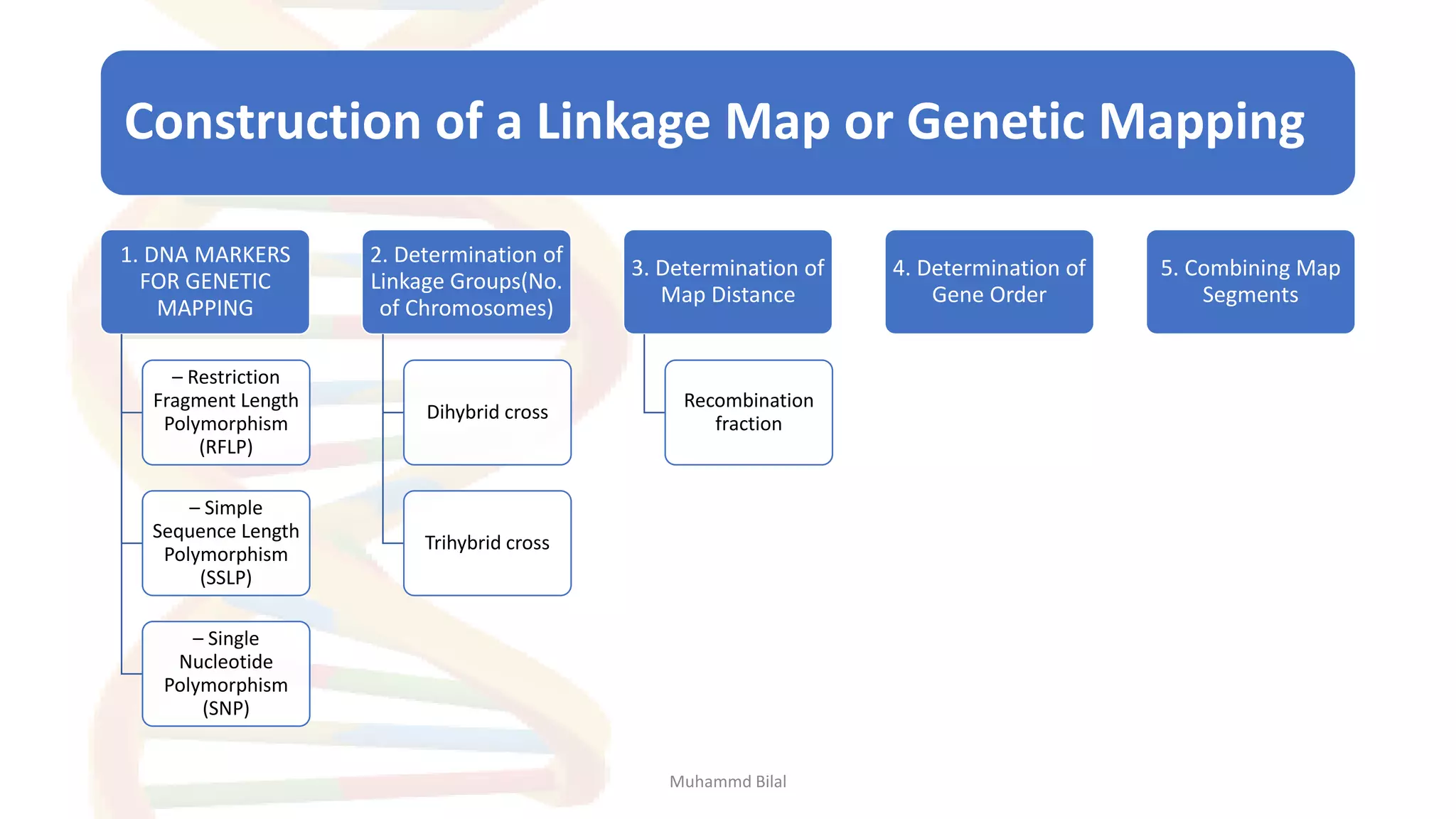
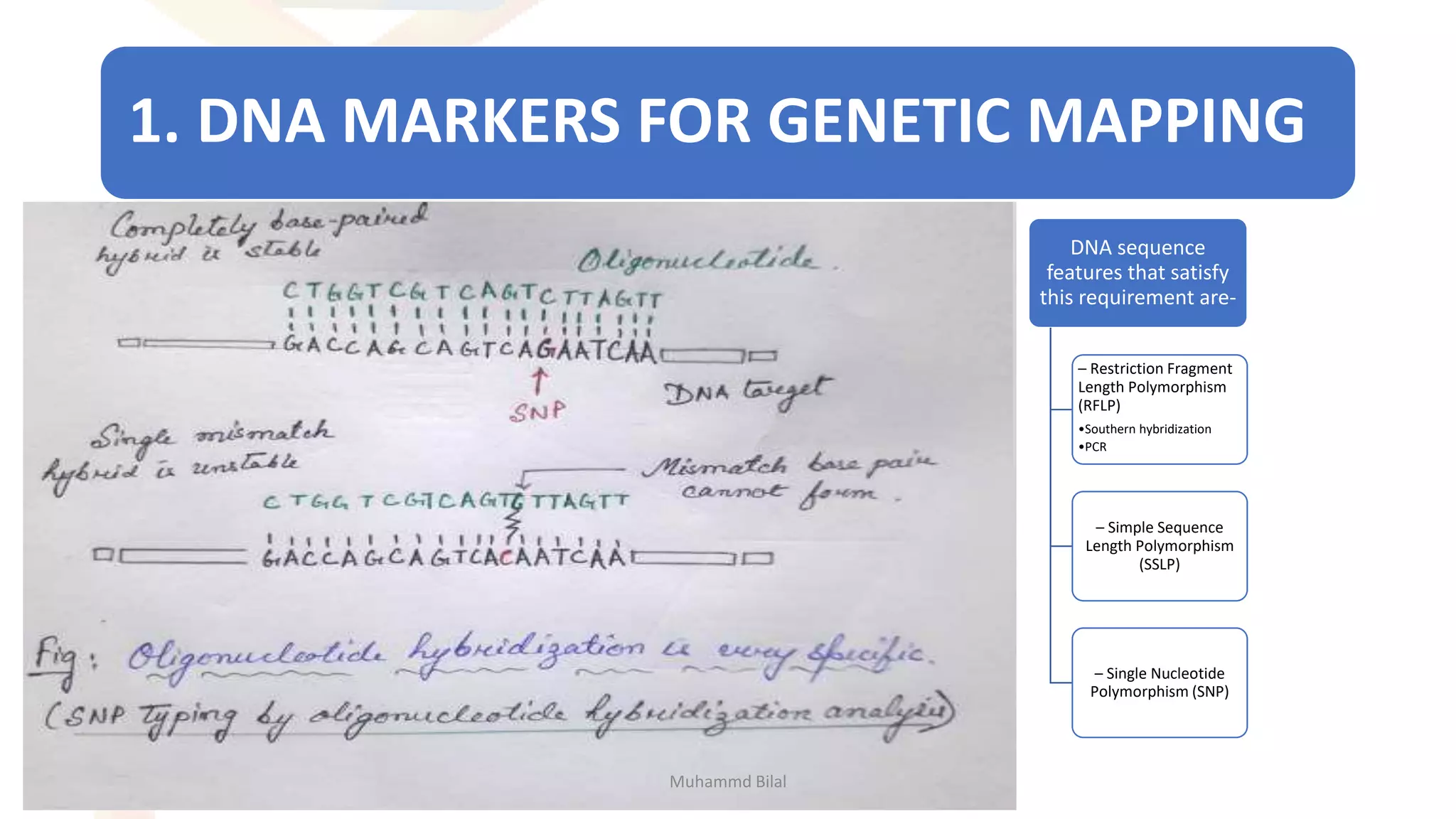
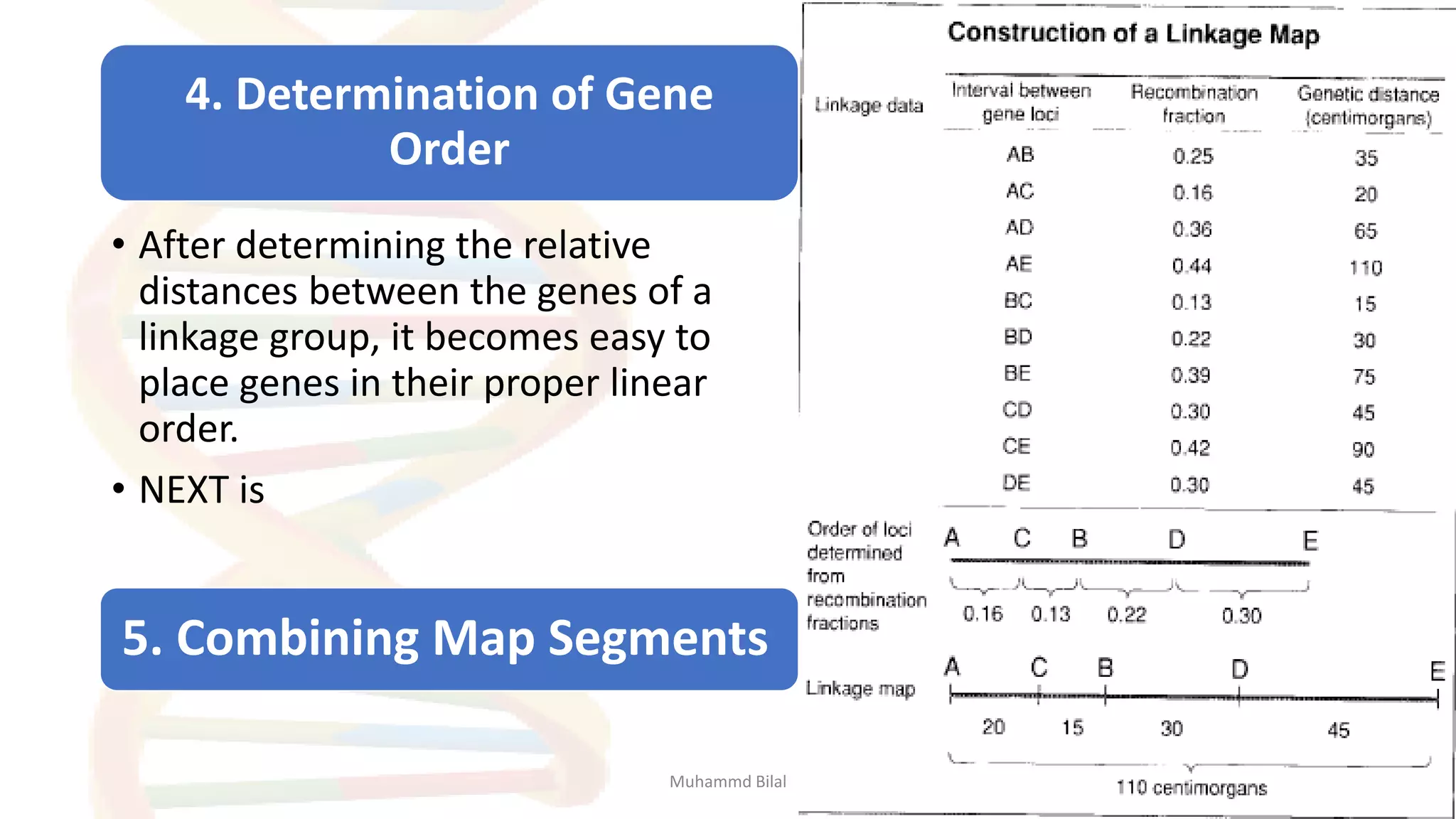
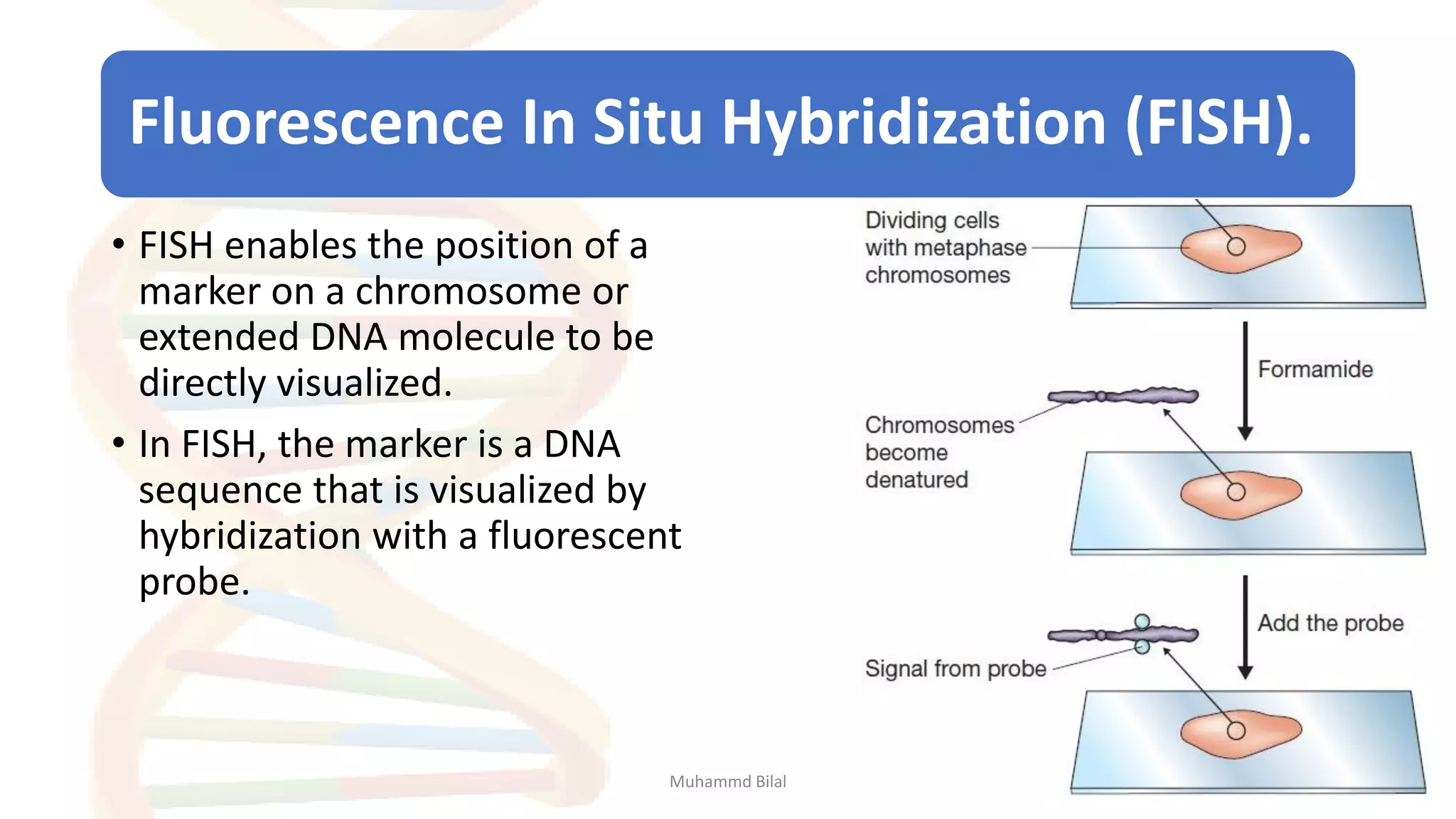
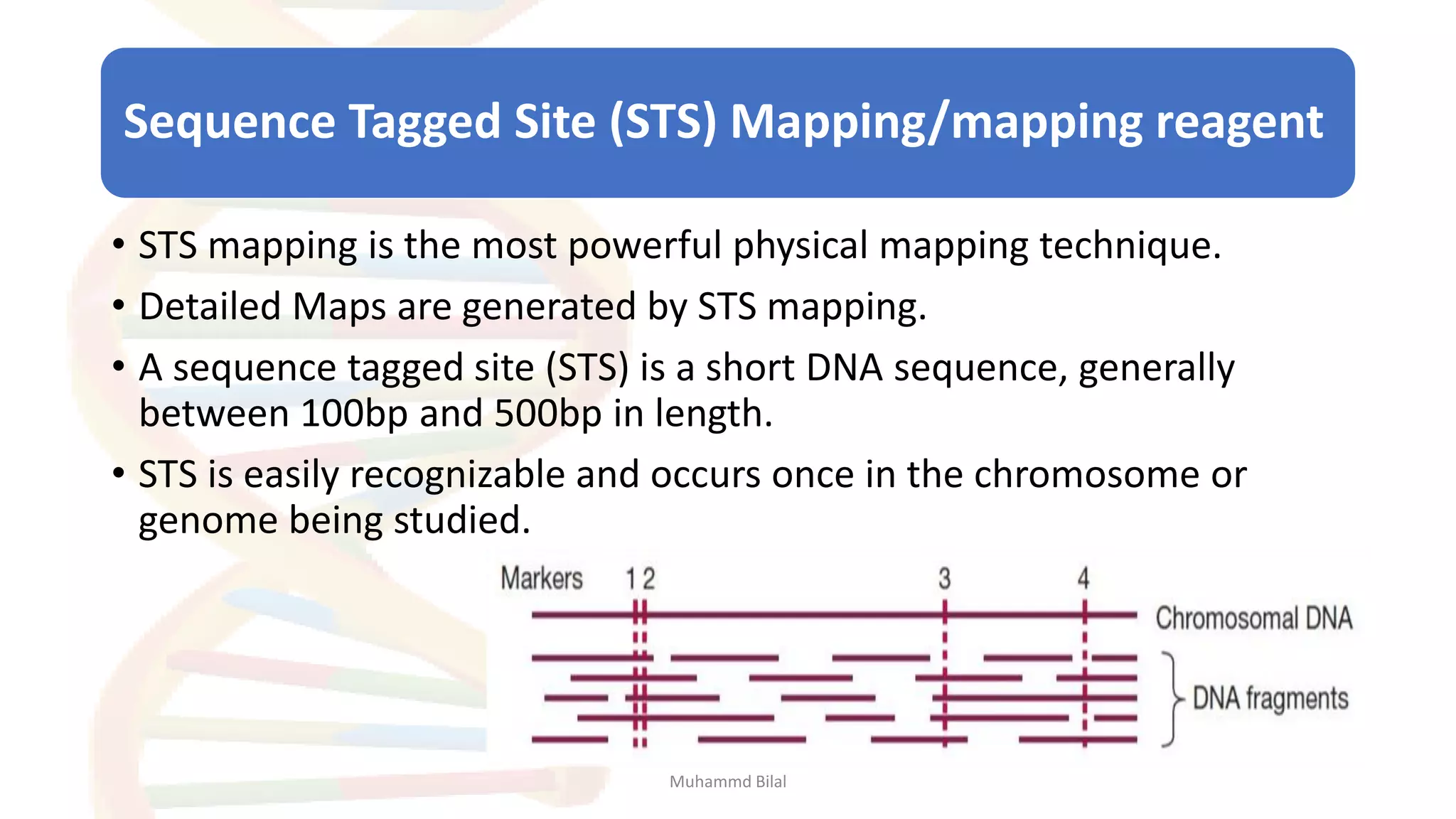
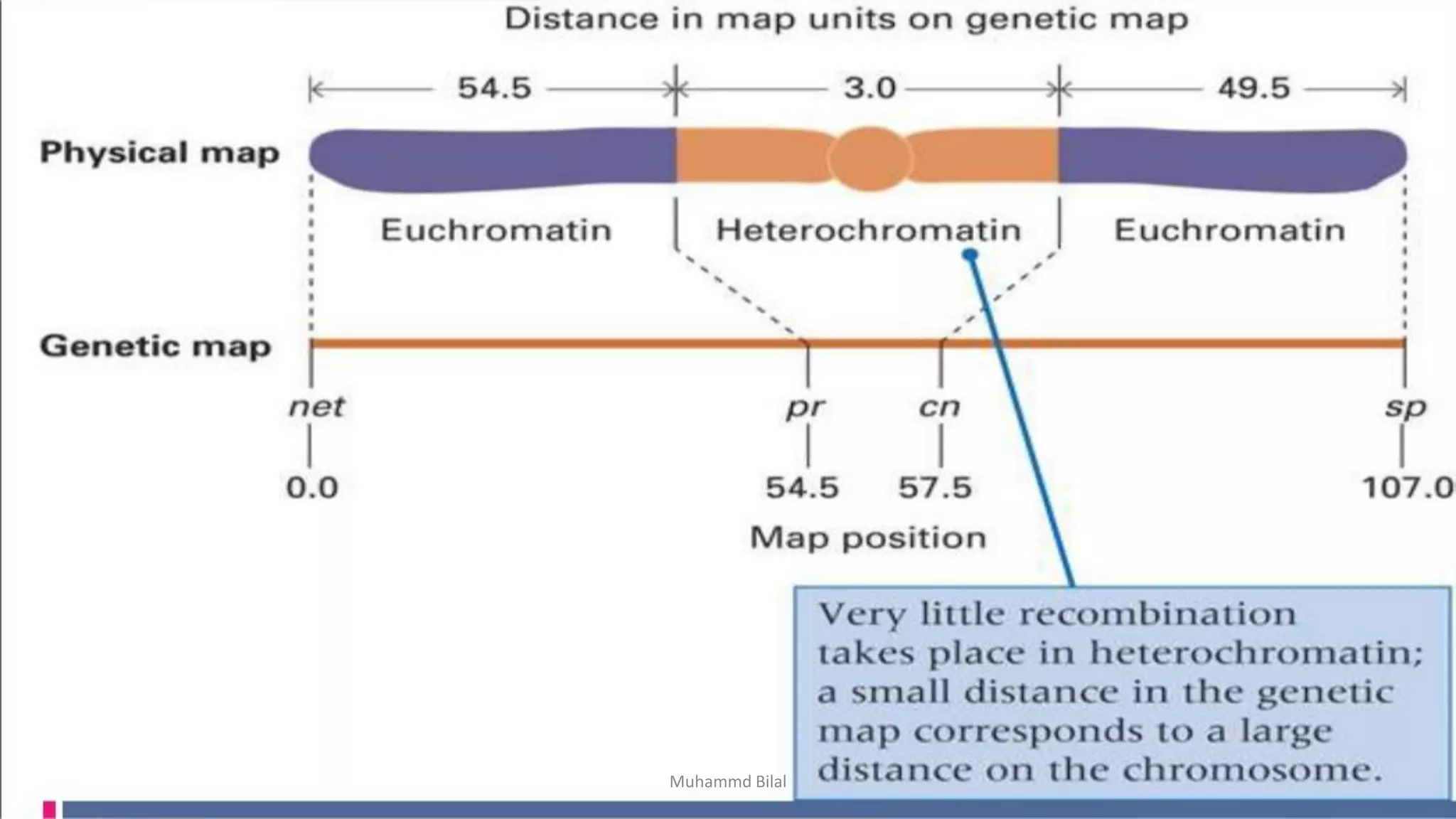
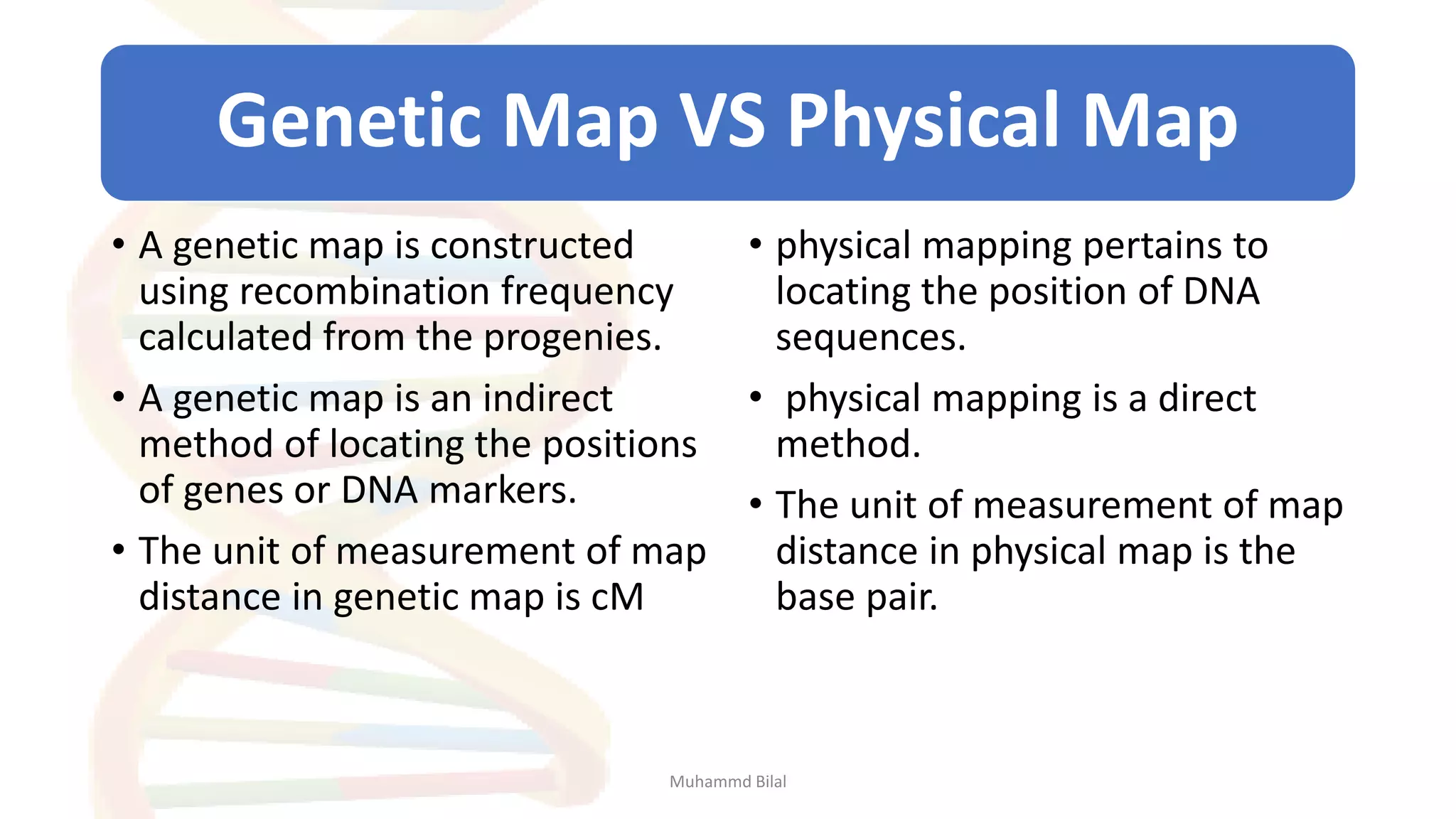
Gene mapping identifies the location and distance of genes within a genome, essential for understanding genetic organization and predicting inheritance patterns. Two primary types include genetic maps, which detail gene distances based on recombination frequency, and physical maps, which directly locate DNA sequences. Despite their utility, genetic maps have limitations in accuracy, leading to the development of physical mapping techniques like fluorescence in situ hybridization (FISH) and sequence tagged site (STS) mapping.