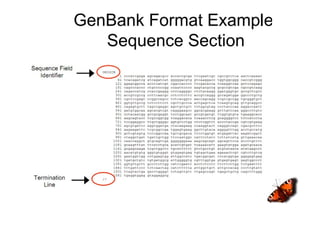
This document discusses several common file formats used for storing bioinformatics sequence data, including their history and key features. It describes early ASCII text file formats, followed by flat file formats adopted for collaboration between sequence databases in 1986 which allowed for annotations. Two common modern formats are then outlined - FASTA format which represents sequences with single letter codes and allows programs to read sequences, and PIR format which identifies sequence type and includes the sequence itself along with optional descriptions. The document also briefly discusses ALN/ClustalW and GCG/MSF formats for storing aligned multiple sequences.