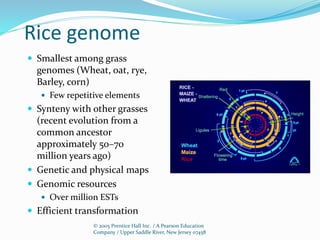
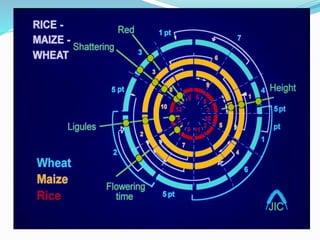
Cereals such as rice, wheat, maize, and barley are economically important crop plants. Rice was chosen as the first cereal genome to sequence due to its small genome size and importance as a food crop. The sequencing of the rice genome established it as a model for studying cereal genomes. Comparative genomics using rice and other sequenced cereal genomes can provide insights into crop improvement and maintaining high quality crops, with significant impacts on global quality of life.