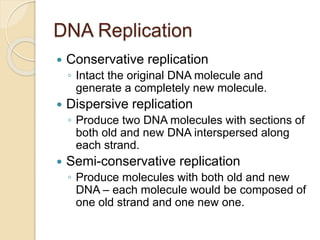
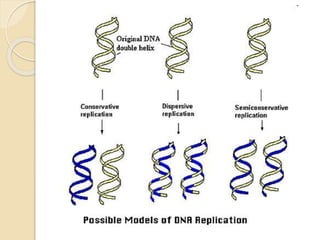
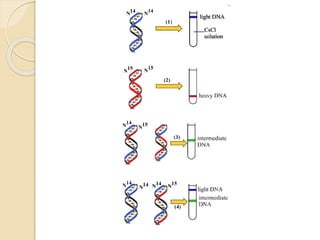
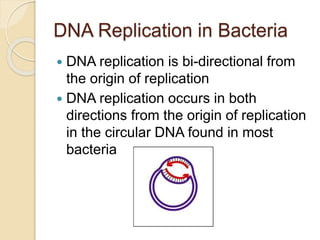
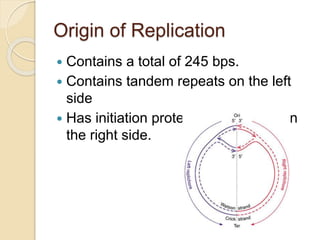
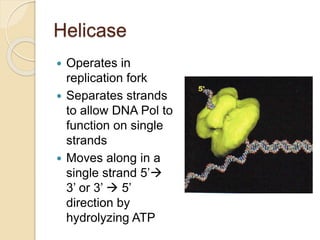
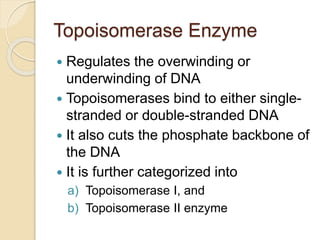
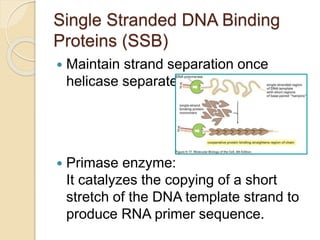
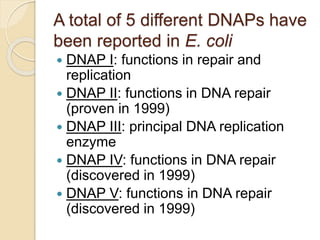
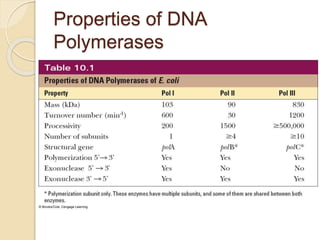
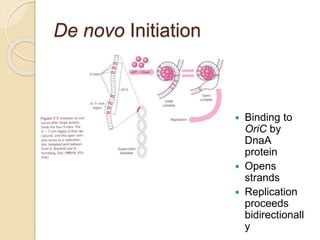
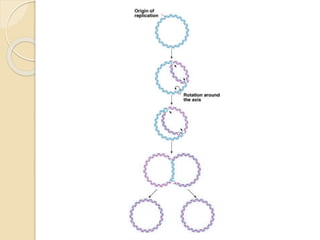
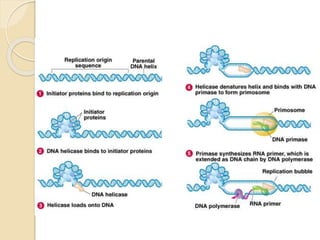
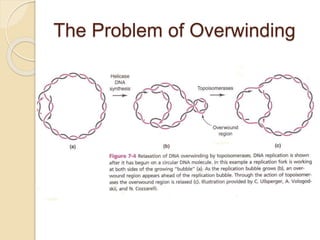
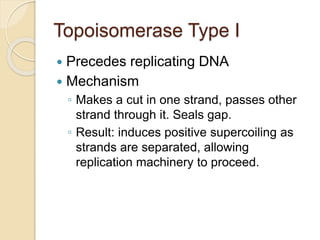
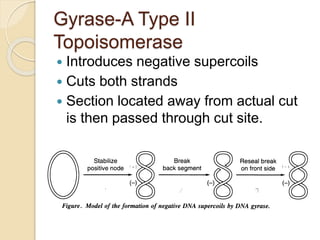
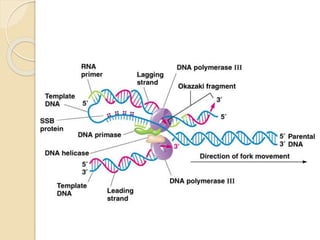
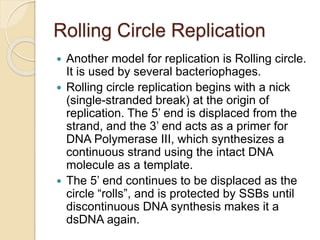
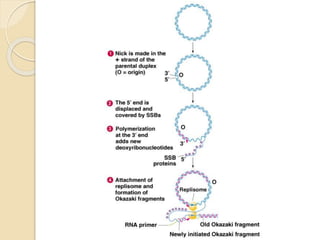
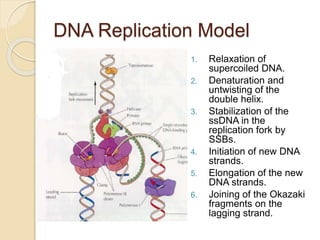
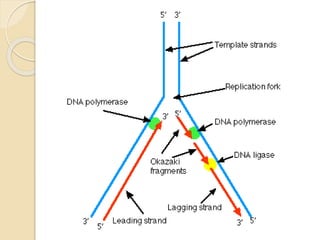
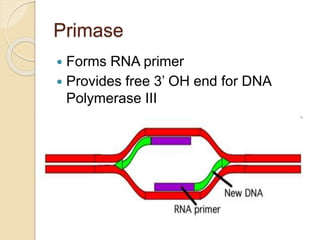
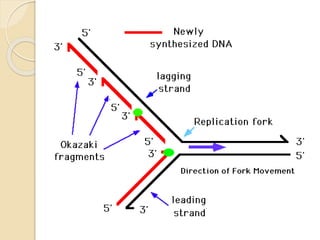
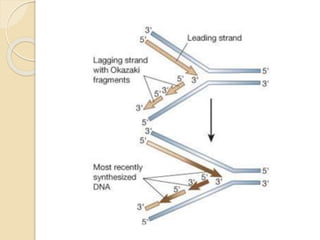
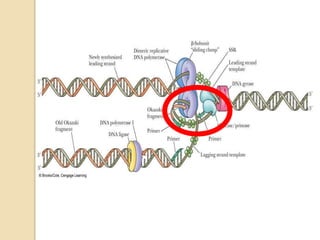
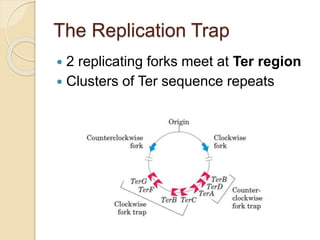
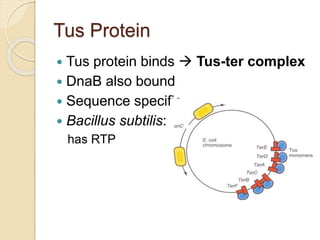
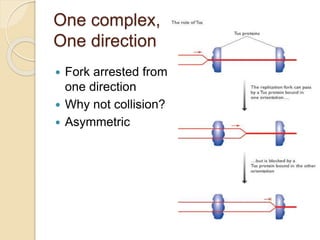
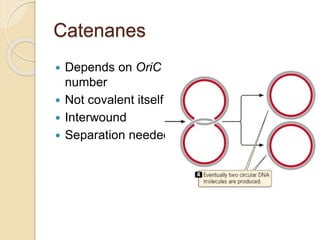
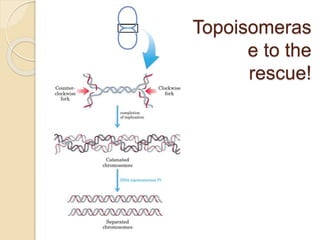
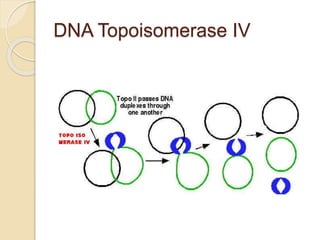
The document summarizes key aspects of DNA replication in bacteria. It describes the semi-conservative model of replication, which was experimentally proven by Meselson and Stahl using E. coli. Replication initiates at a fixed origin and proceeds bidirectionally. It involves unwinding of DNA by helicase, synthesis of new strands by DNA polymerase, and ligation of Okazaki fragments. Replication is tightly regulated to occur once per cell cycle.