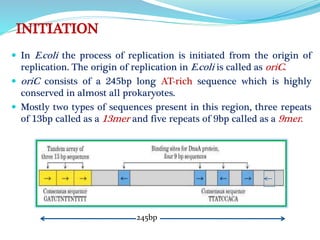
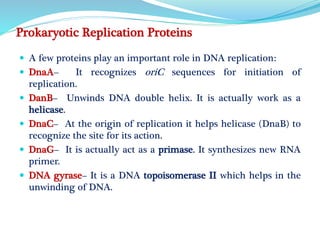
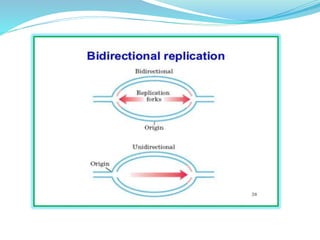
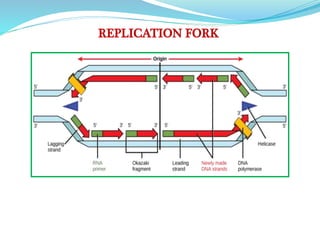
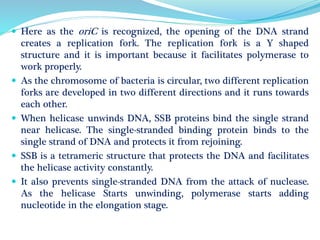
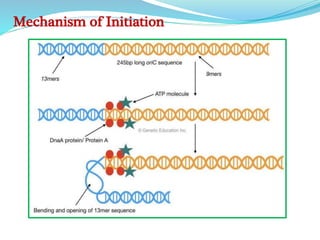
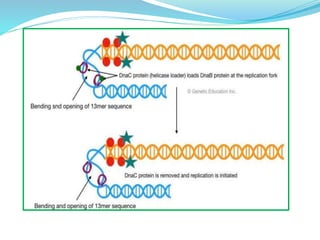
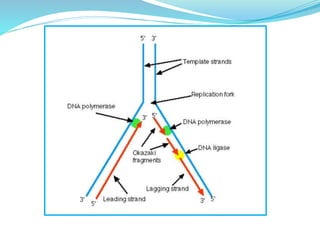
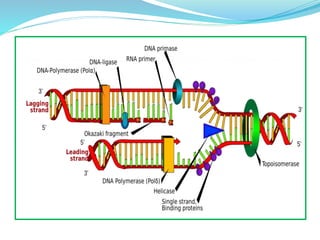
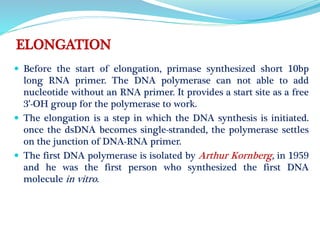
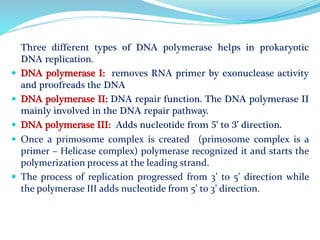
The document summarizes the process of DNA replication in prokaryotes. It describes that replication initiates at the origin of replication (oriC) site and proceeds bidirectionally. There are three main steps - initiation, elongation, and termination. In initiation, proteins help unwind DNA at oriC. In elongation, primase synthesizes primers and DNA polymerase adds nucleotides to replicate both leading and lagging strands. In termination, RNA primers are removed and DNA ligase seals the replicated DNA, completing replication.