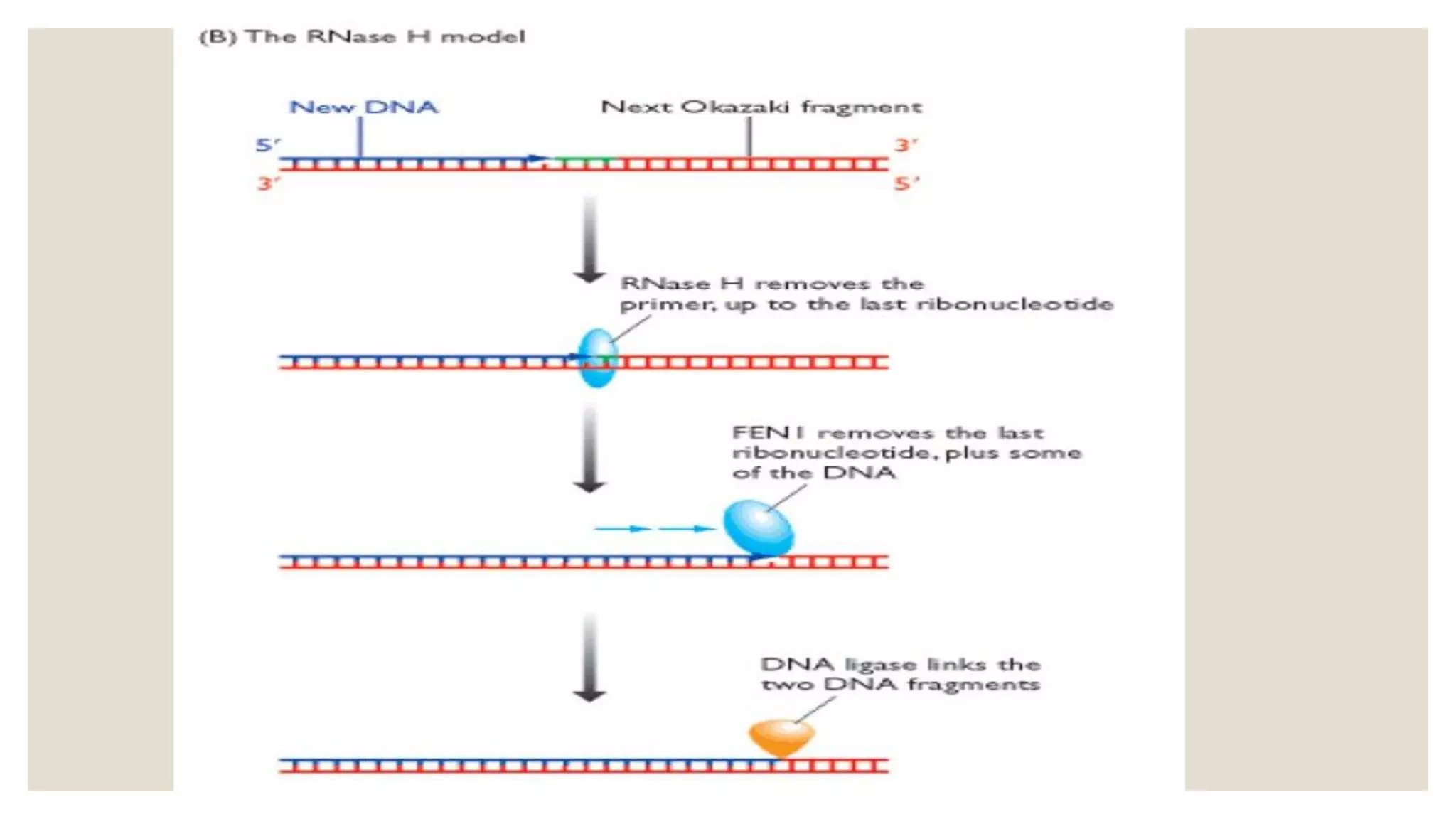
DNA replication in eukaryotes is a complex process that involves multiple origins of replication, mainly characterized by leading and lagging strands, and occurs in three phases: initiation, elongation, and termination. Key proteins such as DNA polymerases α and δ play critical roles in synthesizing new DNA strands, while factors like the autonomous replication sequence (ARS) guide the process. Unique features such as telomeres and the activity of flap endonuclease-1 (Fen-1) also contribute to the fidelity and completeness of DNA replication.