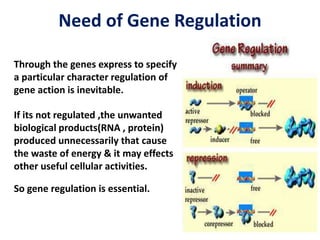
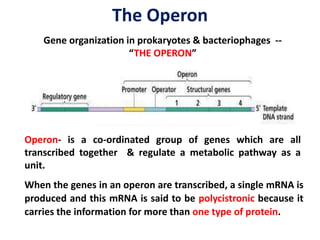
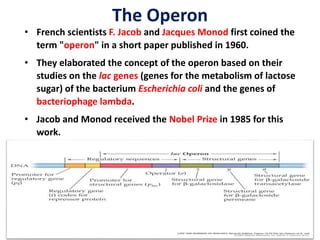
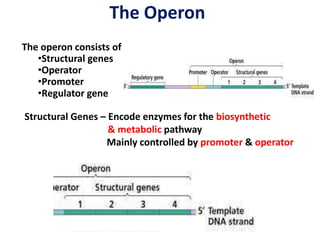
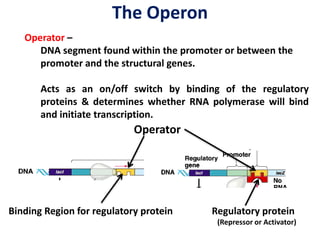
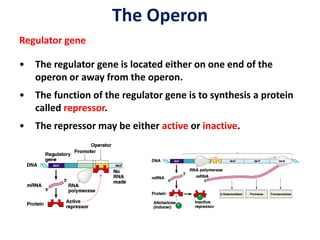
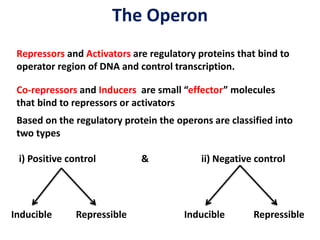
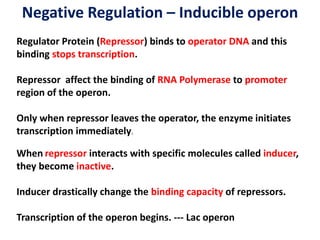
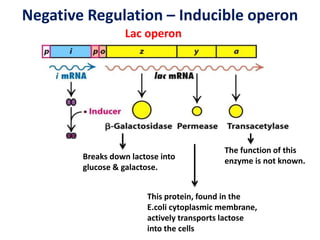
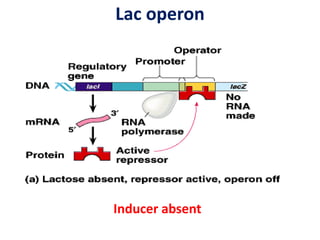
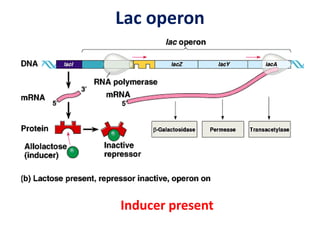
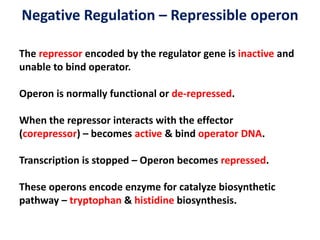
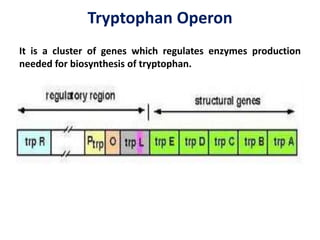
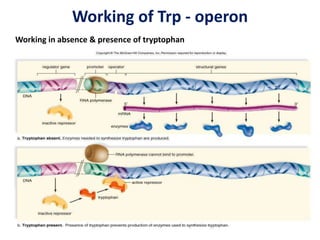
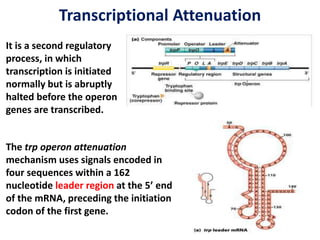
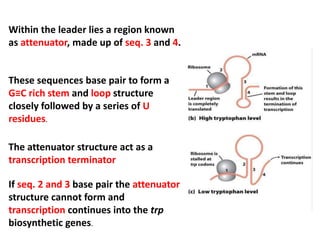
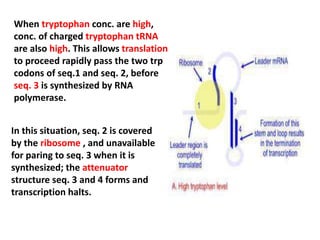
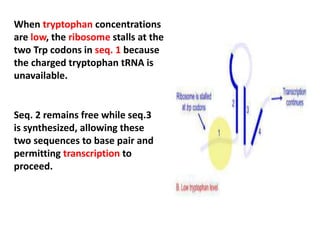
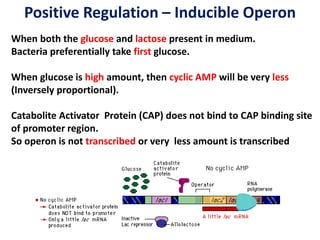
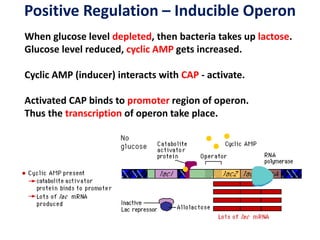
The document discusses the concept of operons in prokaryotes. It defines an operon as a coordinated group of genes that are transcribed together to regulate a metabolic pathway. The key components of an operon are structural genes, an operator, promoter and regulator gene. Operons can be regulated by repressors or activators binding to the operator, which determines if transcription occurs. Examples discussed include the lac and tryptophan operons, which are regulated by negative and positive control, respectively. Transcriptional attenuation is also described as another level of gene regulation. In conclusion, precise regulation of gene expression is essential for organisms to produce the correct phenotypes under different conditions.