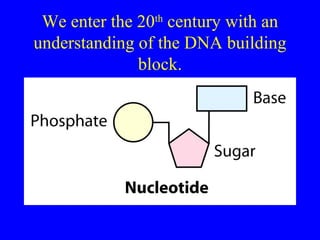
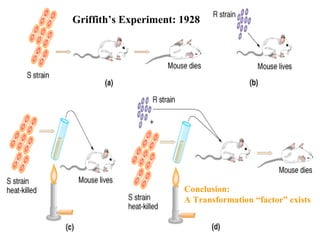
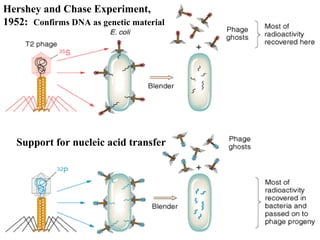
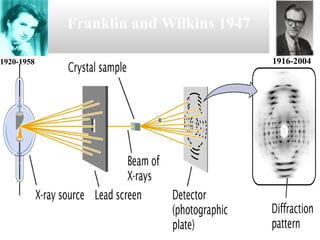
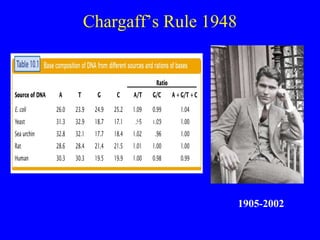
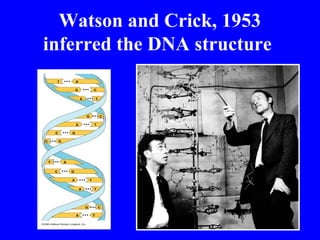
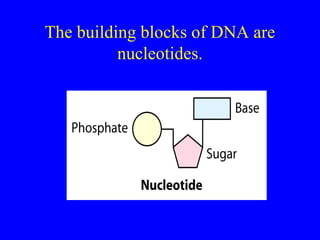
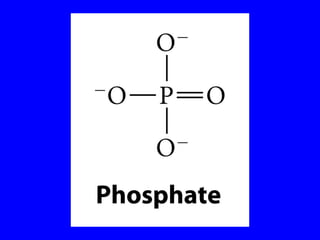
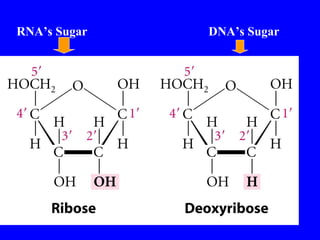
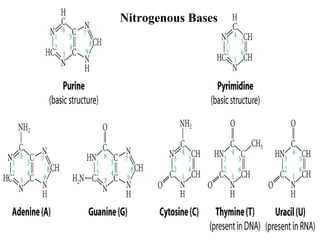
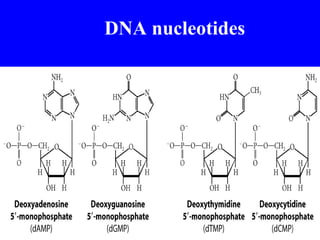
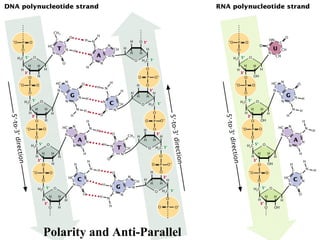
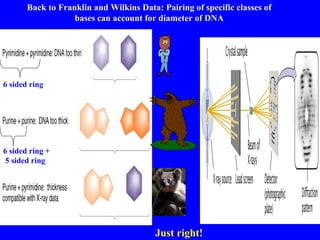
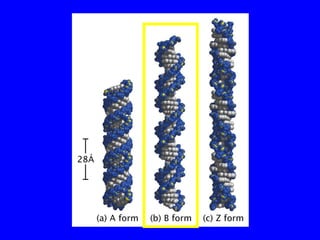
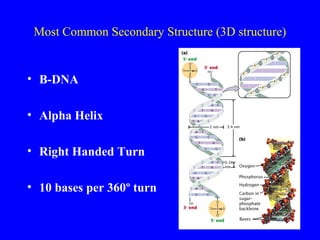
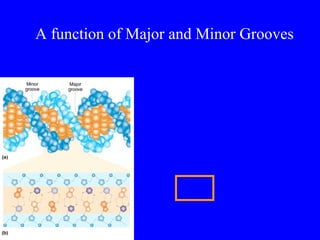
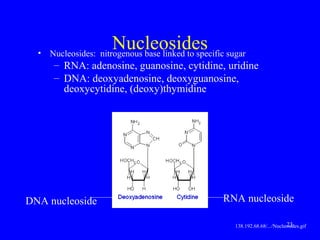
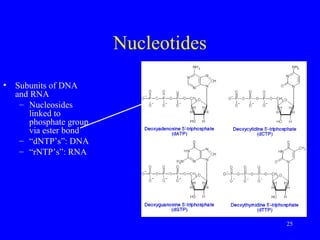
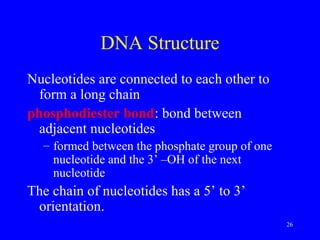
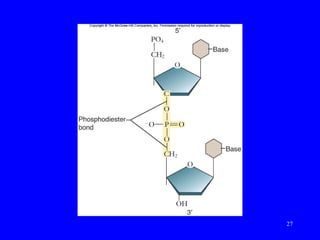
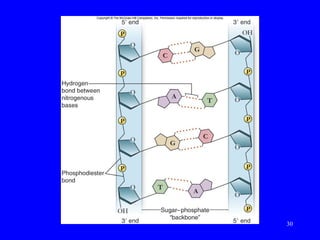
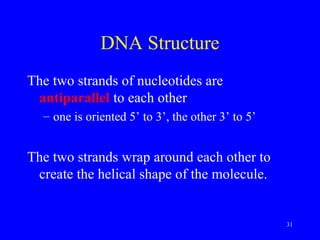
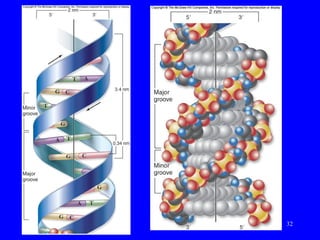
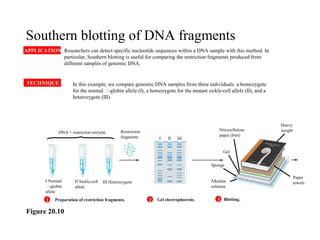
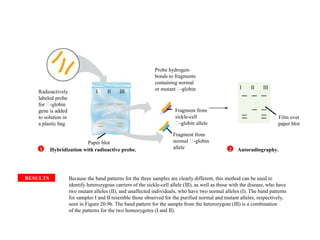
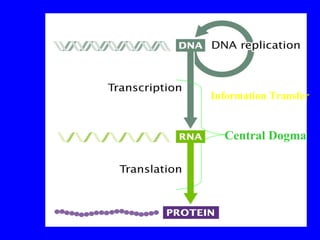
The document provides a summary of the history and discovery of DNA. It discusses early experiments in the 1920s-1950s that provided evidence DNA was the genetic material, including Griffith's experiment, Avery's work, and Hershey and Chase's experiment. It then summarizes Watson and Crick's pivotal discovery in 1953 of the double helix structure of DNA, for which they received the Nobel Prize in 1962. The document outlines the chemical and physical properties of DNA including its nucleotides, base pairing, and double helix structure.