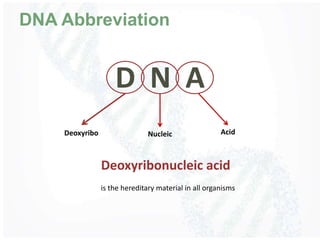
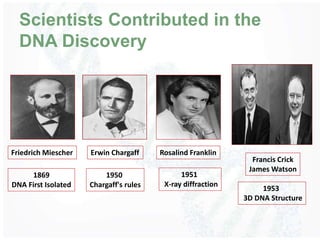
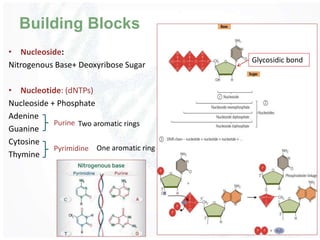
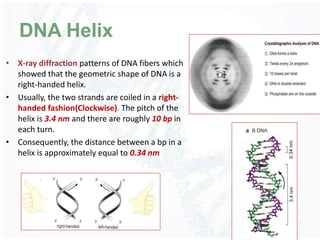
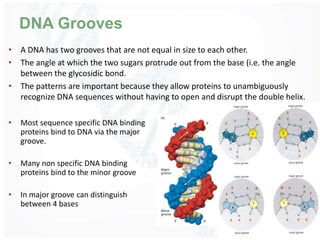
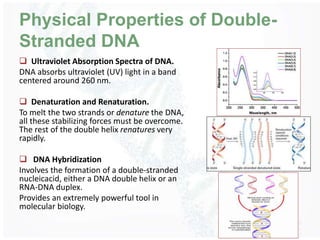
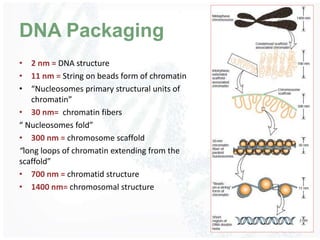
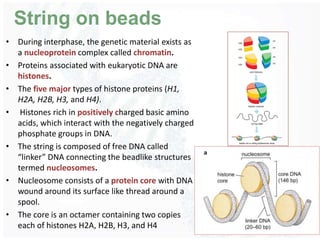
The document provides an overview of DNA structure, including its historical discovery, chemical composition, and the double helix formation. It discusses the various forms of DNA, its packaging into chromosomes, and key properties such as denaturation and hybridization, along with the differences between prokaryotic and eukaryotic DNA. Additionally, it highlights the significance of DNA's structure in gene expression and overall organism complexity.