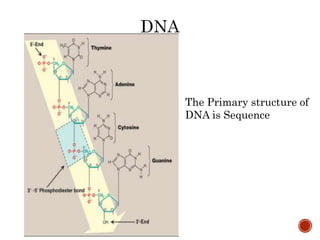
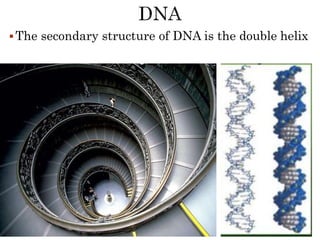
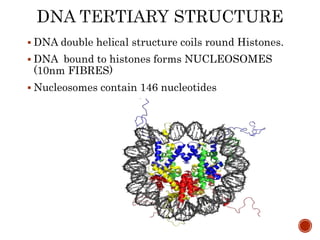
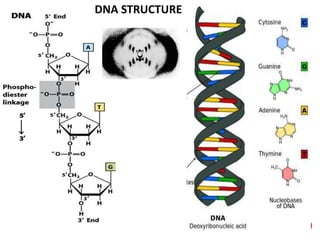
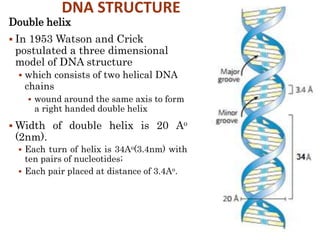
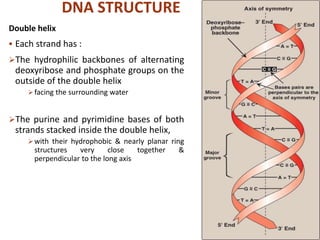
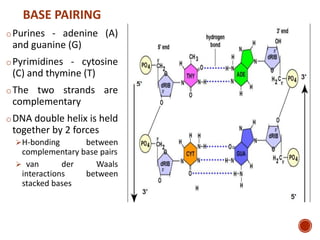
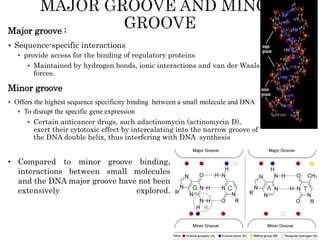
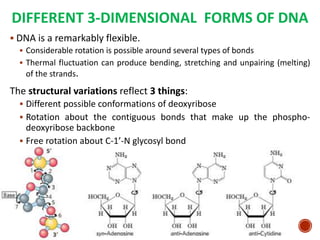
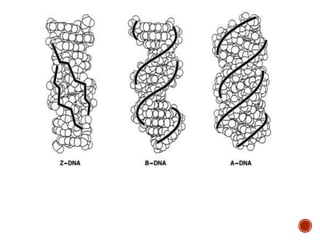
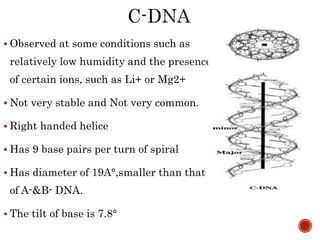
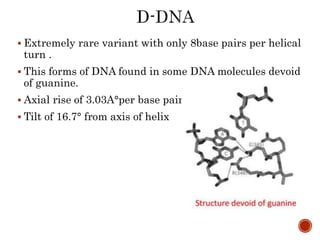
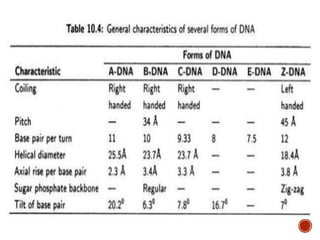
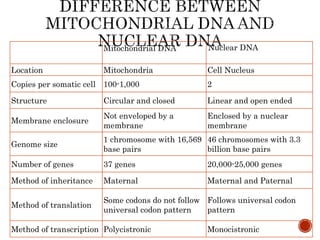
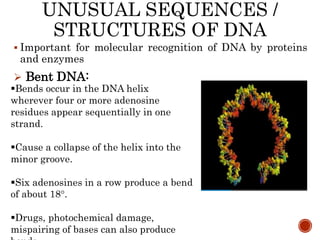
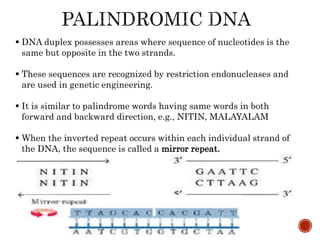
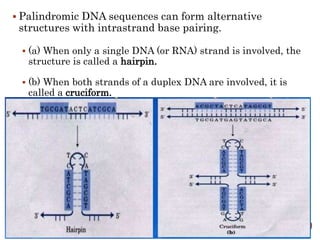
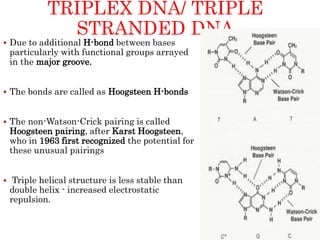
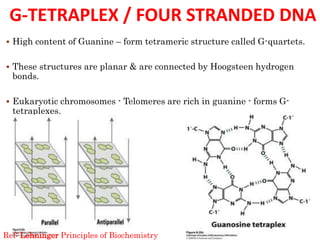
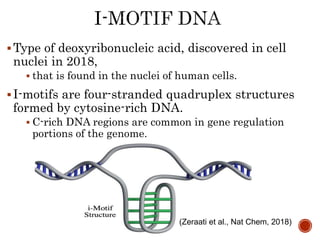
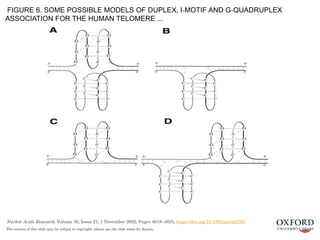
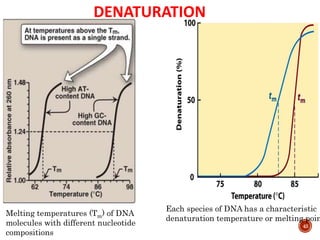
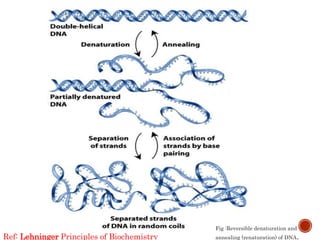
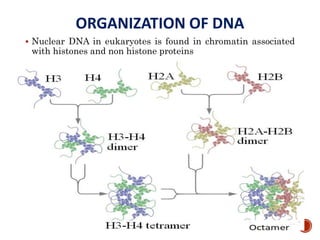
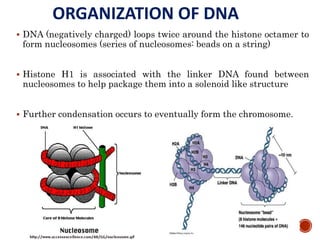
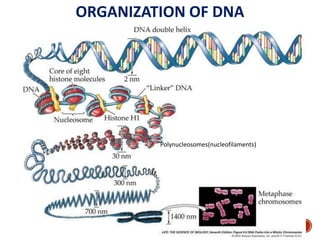
The document provides a comprehensive overview of DNA, detailing its history, structure, functions, types, and implications in genetics. Key milestones include the identification of DNA by Friedrich Miescher and the elucidation of its double helix structure by Watson and Crick, with descriptions of various forms of DNA and their biological roles. Additionally, it discusses genetic information, replication, mutations, transcription, and the significance of DNA in areas such as gene therapy and forensic science.