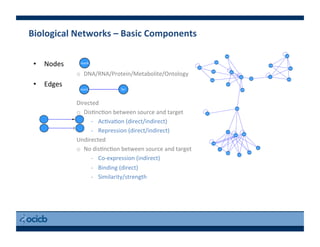
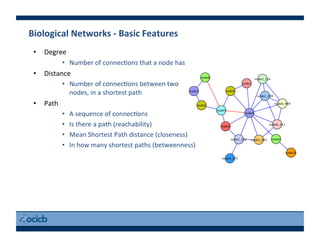
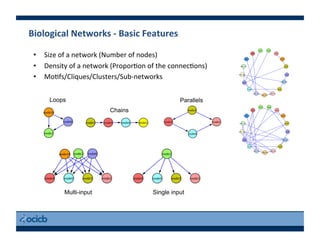
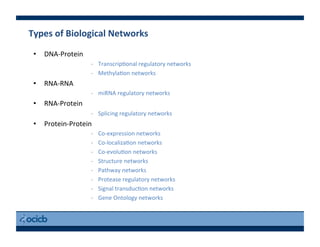
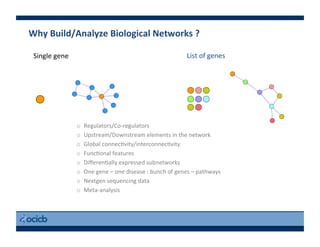
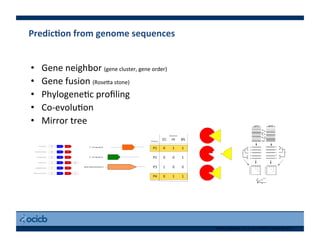
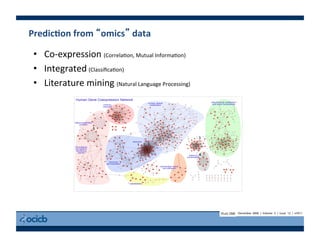
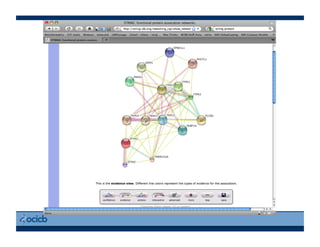
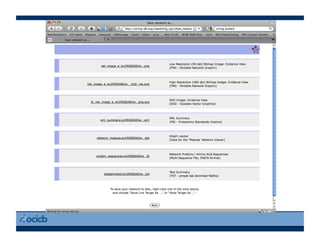
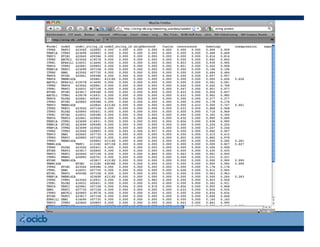
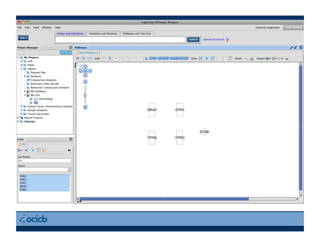
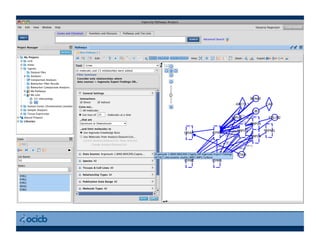
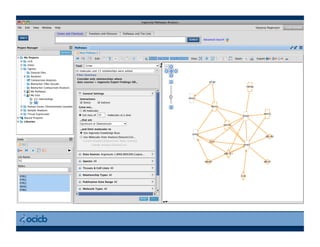
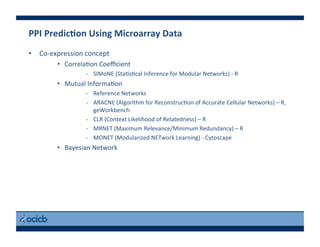
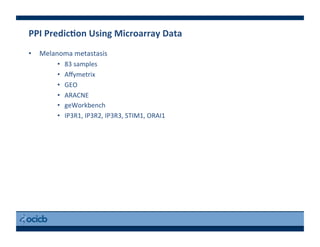
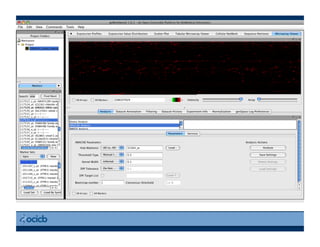
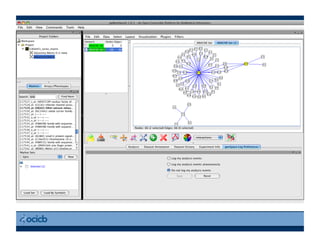
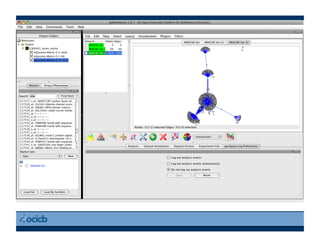
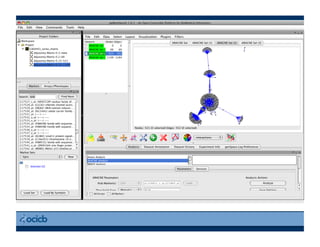
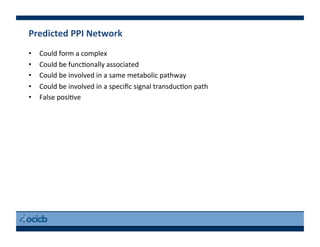
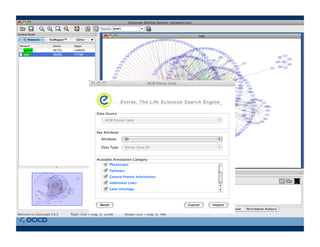
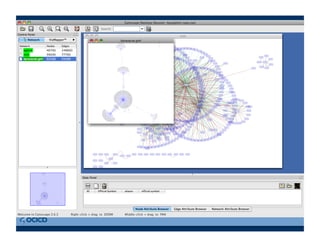
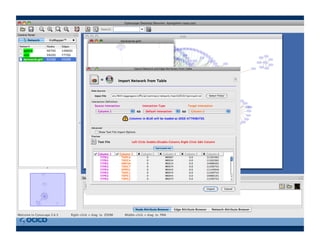
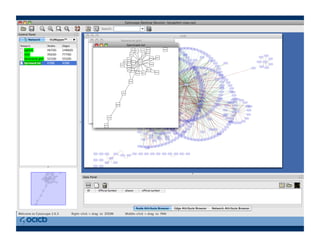
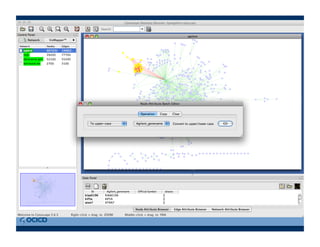
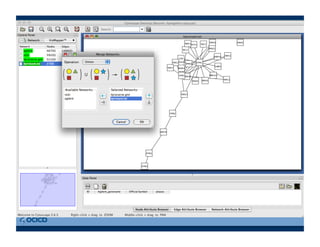
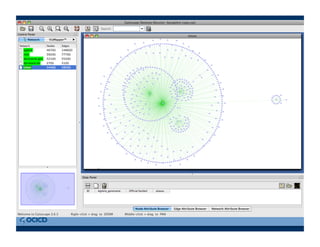
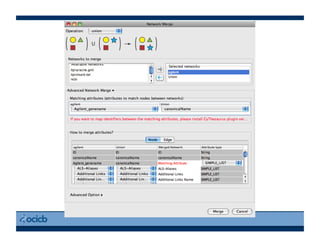
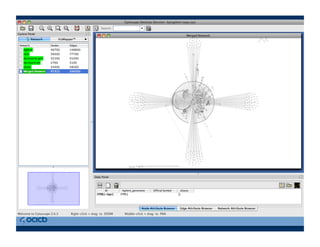
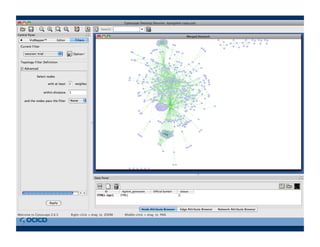
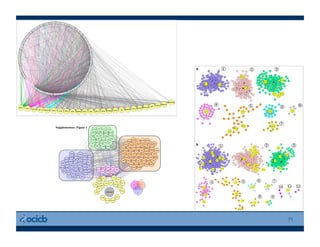
This document provides an outline for a presentation on biological networks, including introducing biological networks, describing their basic components and types, methods for predicting and building networks, sources of interaction data, tools for network visualization and analysis, and a demonstration of building, visualizing and analyzing biological networks using Cytoscape. The presentation covers topics like nodes and edges in networks, features used to analyze networks, methods for predicting networks from sequences and omics data, integrated databases for interaction data, and popular tools for searching, visualizing and performing network analysis.