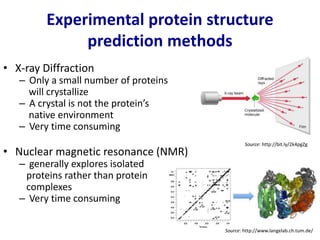
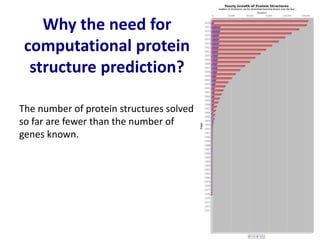
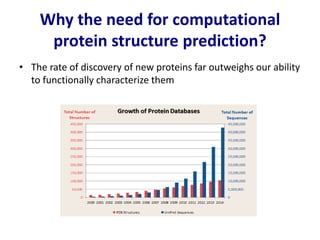
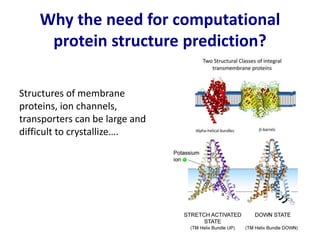
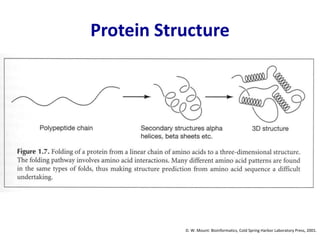
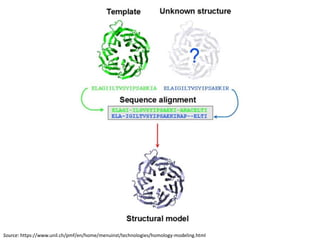
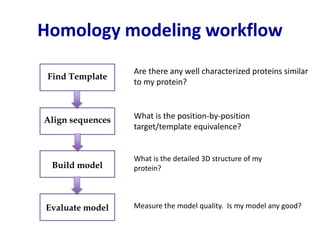
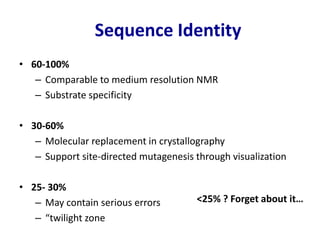
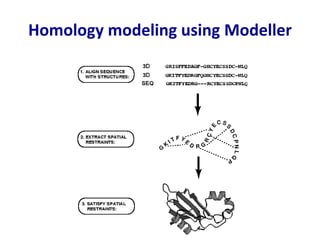
Homology modeling is a computational technique for predicting the structure of a protein target based on its sequence similarity to proteins with known structures, and it involves finding a suitable template, aligning the target and template sequences, building a 3D model of the target, and evaluating the model quality. While experimental methods like X-ray crystallography and NMR can determine protein structures, they have limitations in terms of which proteins can be studied, so computational methods like homology modeling are needed to predict structures for the many proteins whose structures remain unknown.