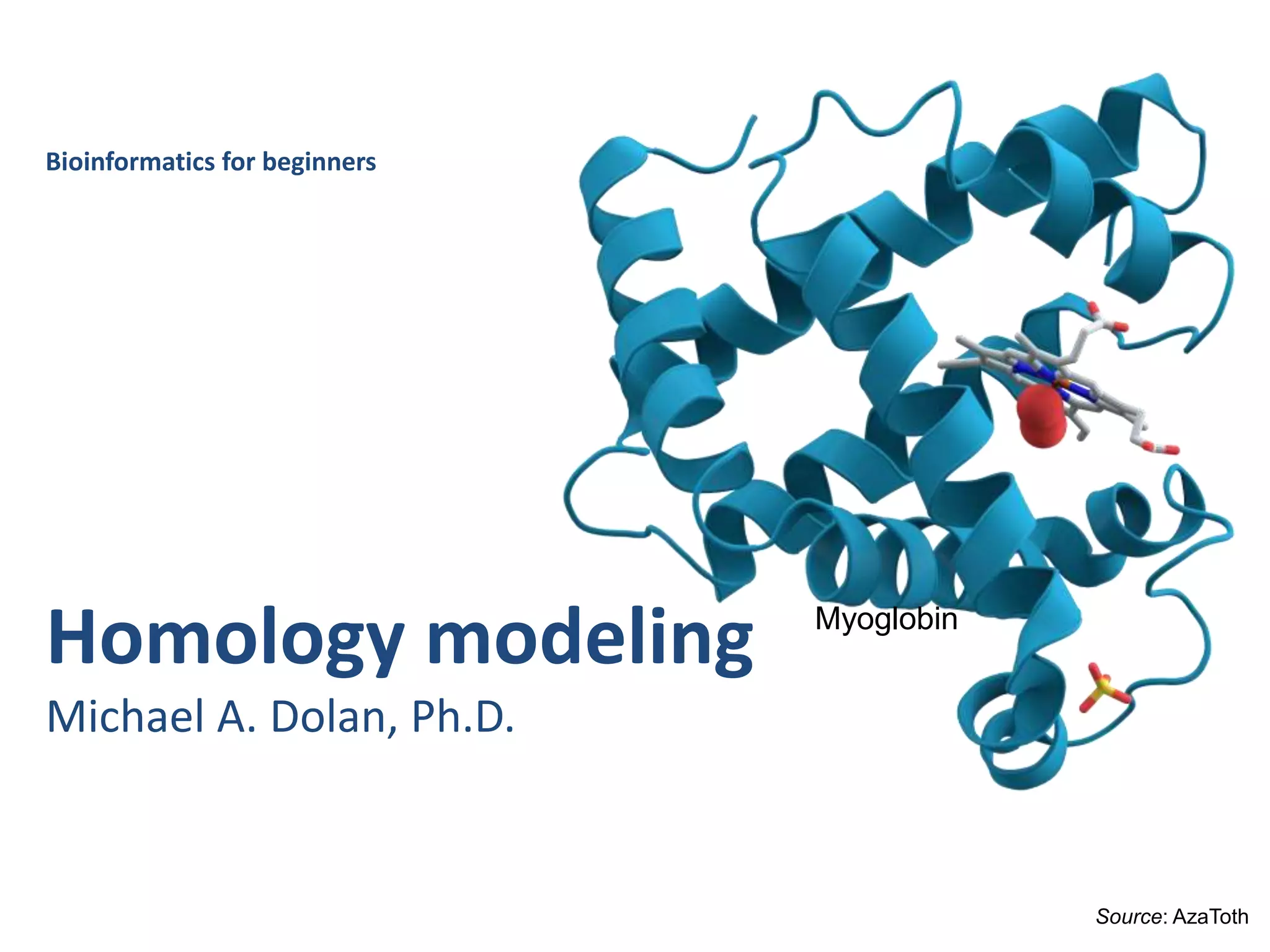
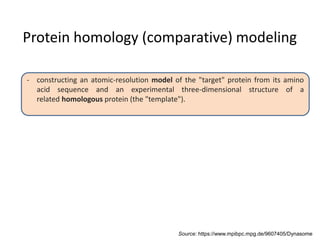
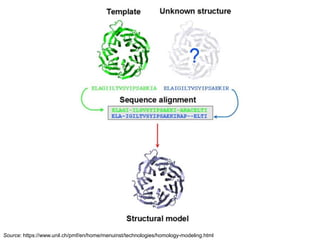
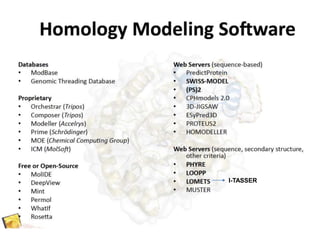
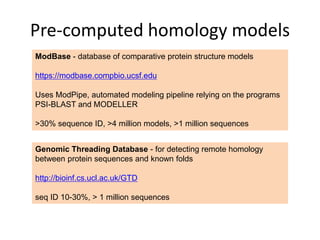
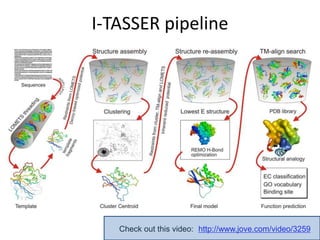
This document provides an introduction to homology modeling using computational tools like I-TASSER and Phyre2. It discusses how homology modeling can be used to generate 3D structural models of proteins when an experimental structure is not available. The document addresses common questions from users and outlines the I-TASSER modeling pipeline. Hands-on exercises are provided to allow users to run homology modeling tools and examine the resulting models.






![Examine the results
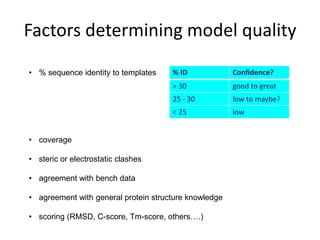
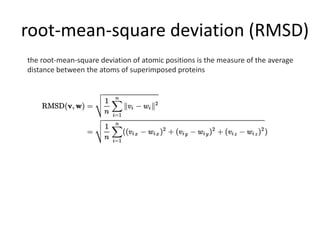
C-score is a confidence score for estimating the quality of models.
• calculated based on the significance of threading template alignments
and the convergence parameters of the structure assembly simulations
• C-score is typically in the range of [-5 to 2], where a C-score of higher
value signifies a model with a high confidence and vice-versa.
Tm-score - solves the problem of local error when calculating RMSD](https://image.slidesharecdn.com/introhomologymodeling-181002220820/85/Intro-to-homology-modeling-15-320.jpg)