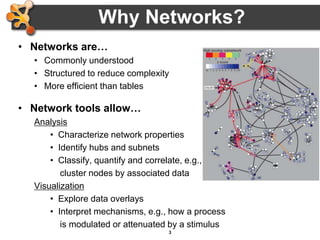
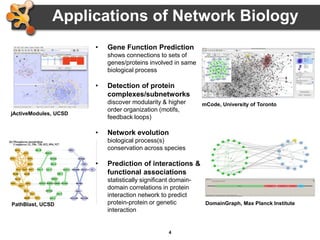
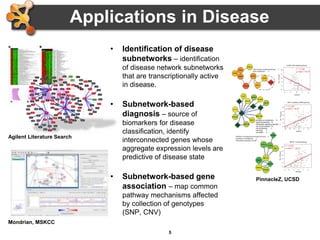
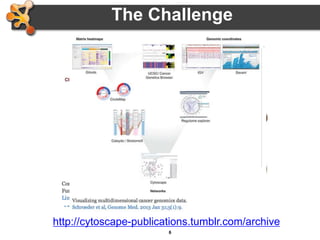
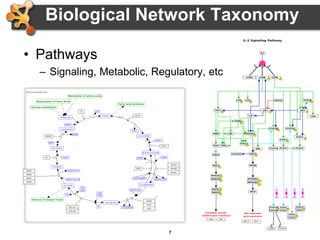
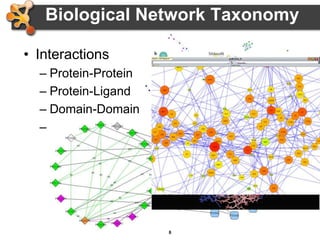
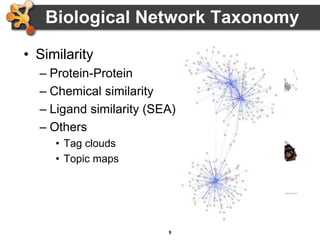
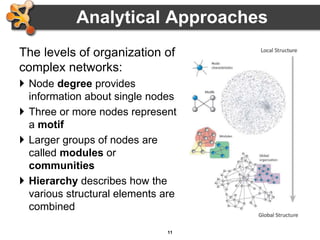
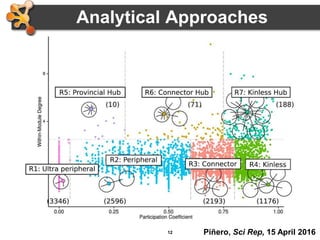
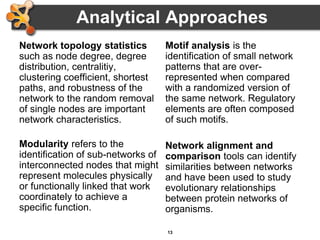
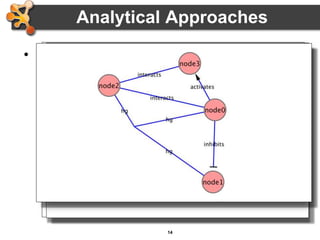
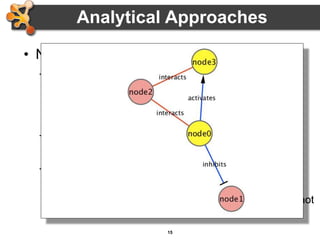
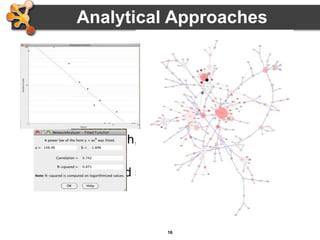
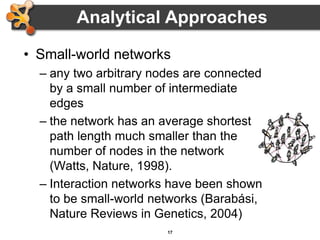
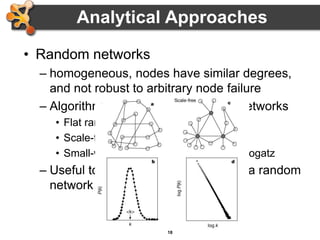
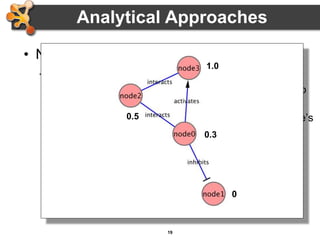
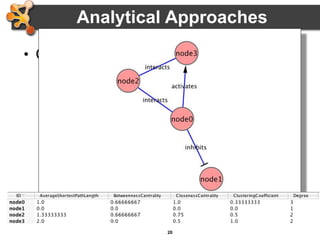
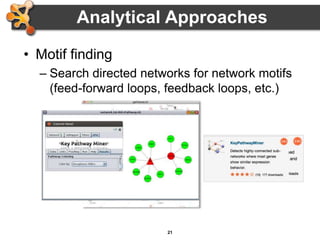
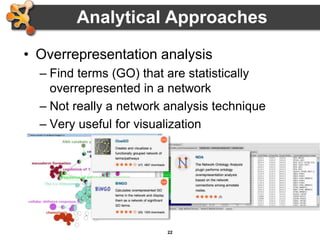
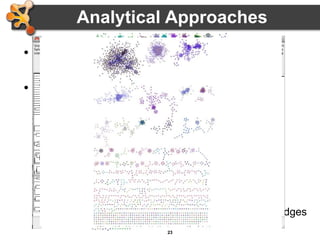
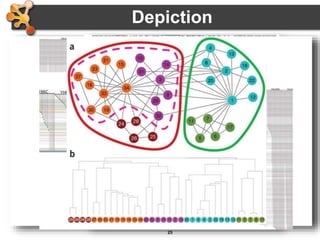
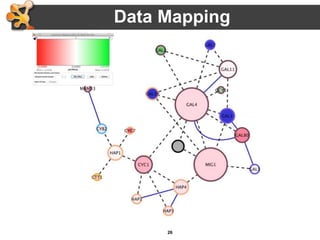
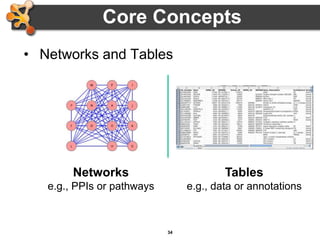
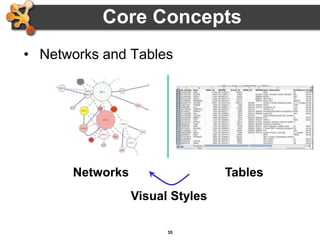
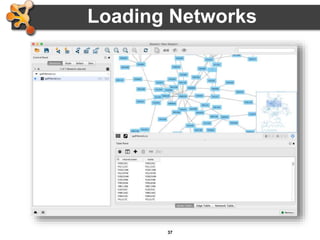
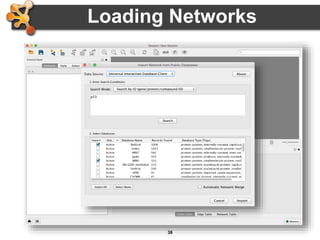
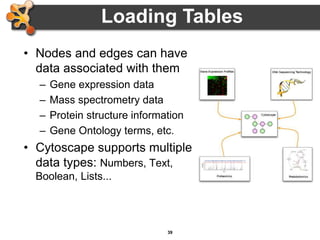
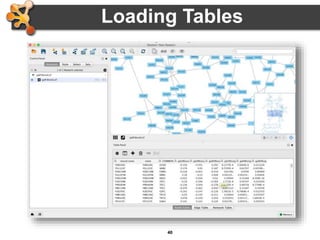
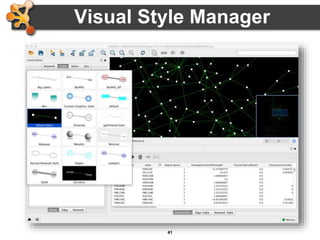
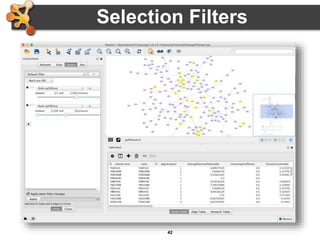
This document outlines a presentation on biological networks and the software Cytoscape. It begins with an introduction to biological networks and their taxonomy, as well as analytical approaches and visualization techniques. It then provides an overview of Cytoscape, covering core concepts like networks and tables, visual properties, and apps. The document demonstrates how to load networks and data, use visual style managers, and save and export networks. It concludes with tips and tricks for using Cytoscape and a link to a hands-on tutorial.