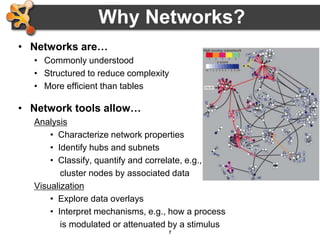
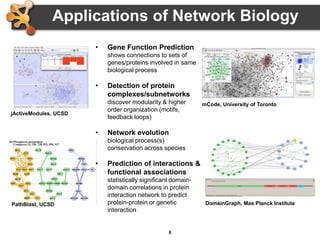
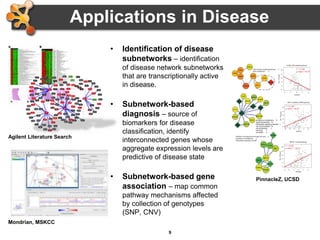
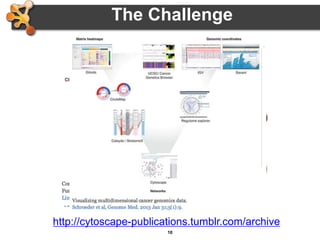
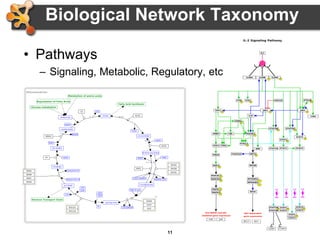
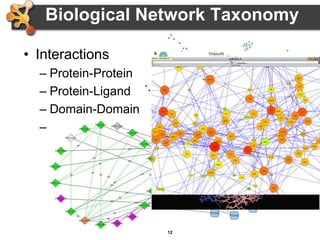
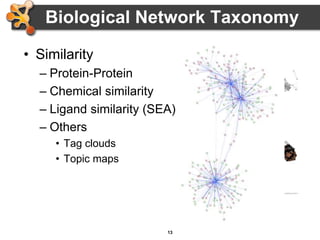
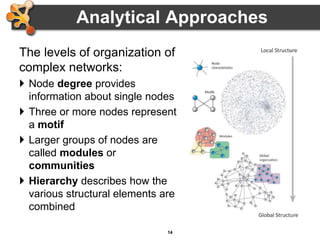
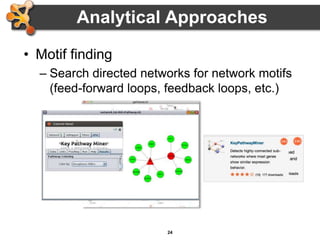
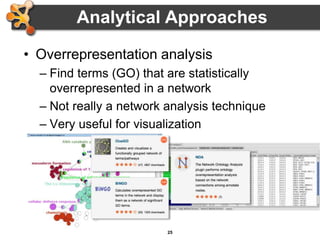
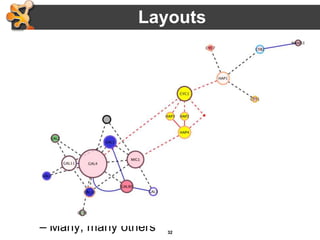
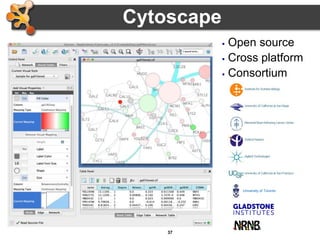
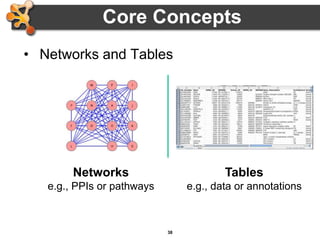
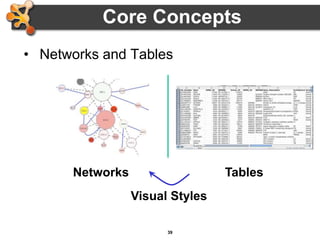
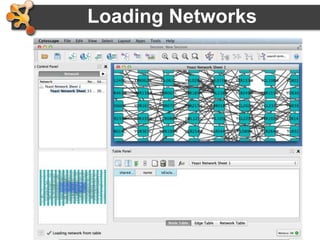
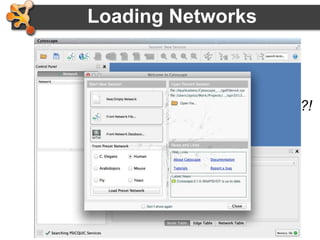
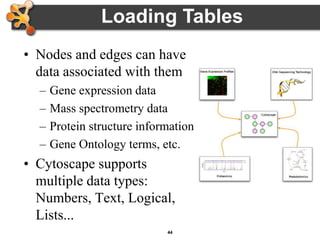
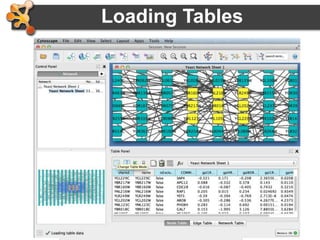
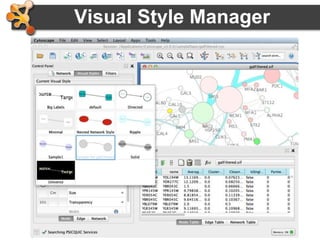
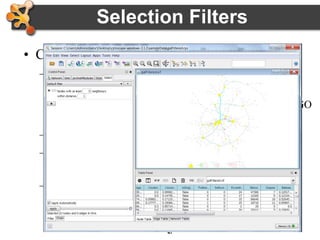
This document provides an introduction and outline for a workshop on using Cytoscape 3 to analyze and visualize biological networks. The workshop will cover core Cytoscape concepts like networks and tables, visual properties, and apps. Participants will learn how to import networks and data, use layouts and apps to explore their own data, and get help resources. The document outlines why networks are important in biology and different analytical approaches and visualization techniques in Cytoscape.