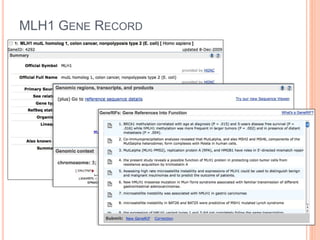
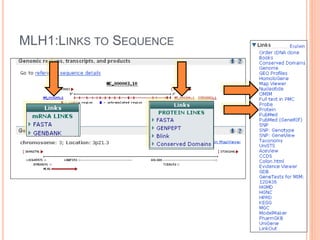
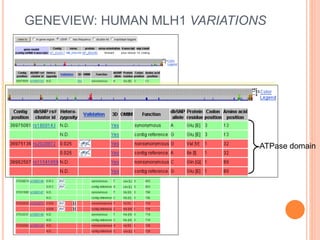
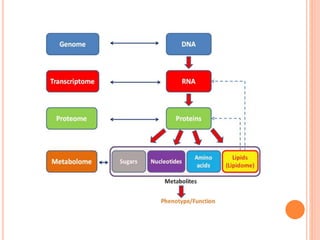
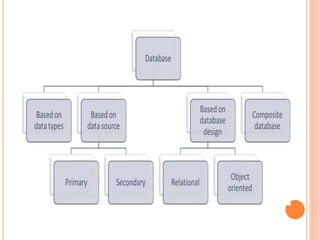
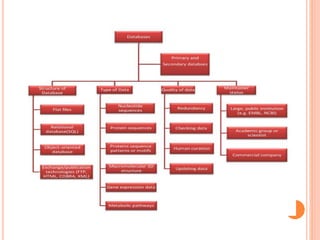
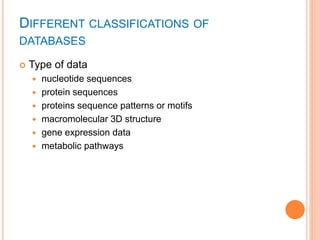
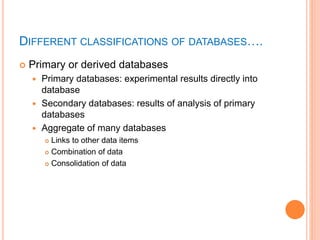
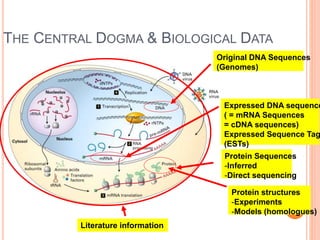
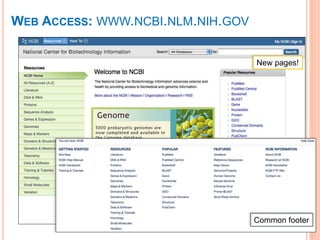
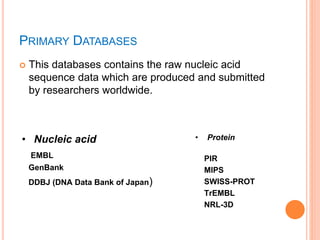
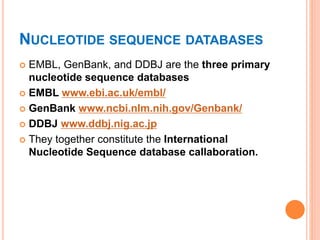
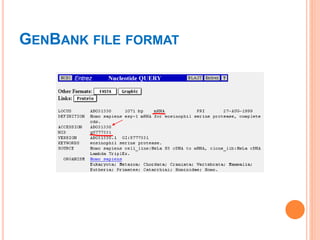
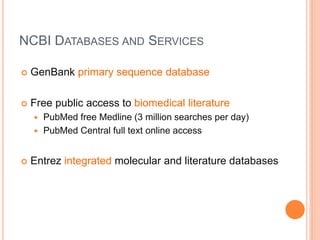
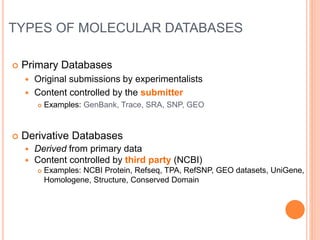
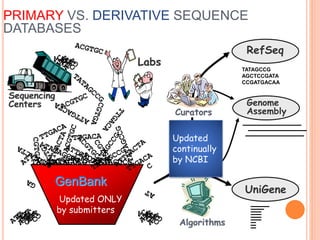
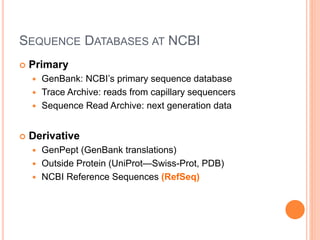
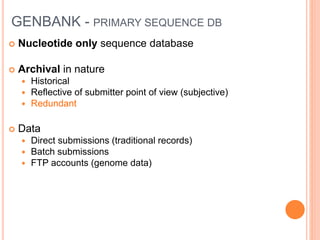
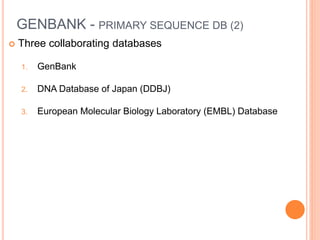
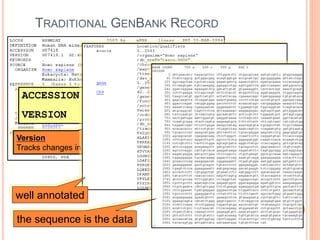
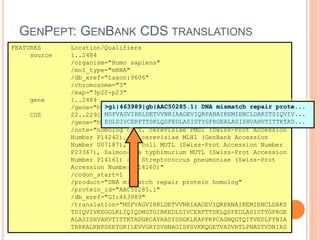
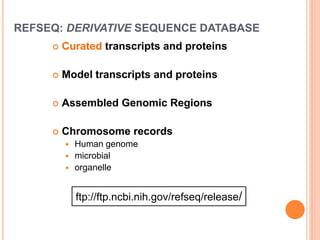
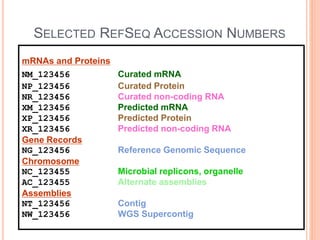
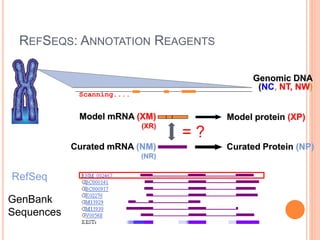
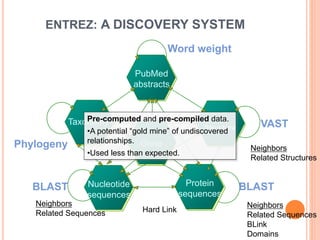
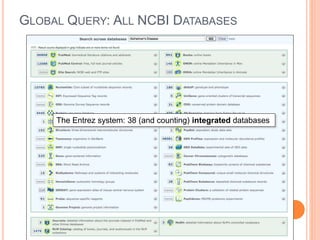
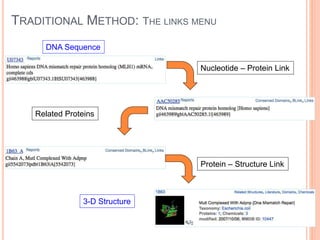
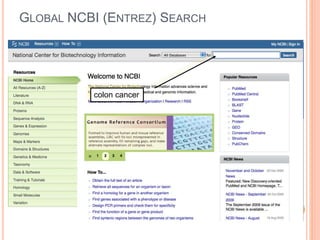
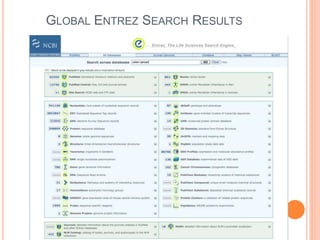
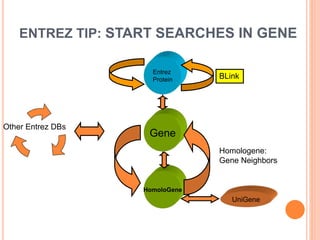
The document discusses biological databases. It begins by defining what a database is, including that it is a collection of related data organized in a way that allows information to be retrieved easily. It then discusses different types of biological databases, including those containing nucleotide sequences, protein sequences, 3D structures, gene expression data, and metabolic pathways. The rest of the document provides details on specific biological databases like GenBank, EMBL, DDBJ, and NCBI databases including Entrez. It emphasizes that biological data is heterogeneous, large in volume, dynamic and integrated across multiple databases.




















![PRECISE RESULTS
MLH1[Gene Name] AND Human[Organism]](https://image.slidesharecdn.com/introductiontodatabases-210511074114/85/Introduction-to-Biological-databases-49-320.jpg)