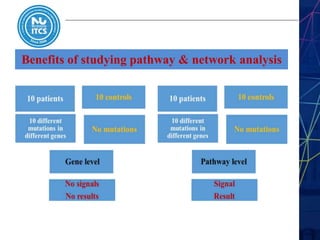
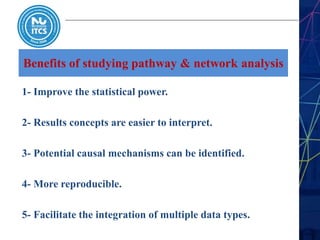
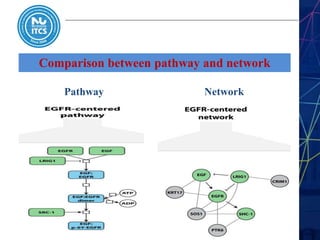
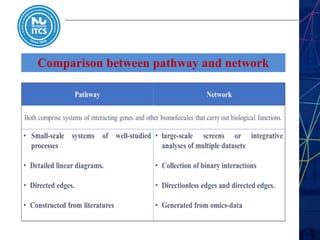
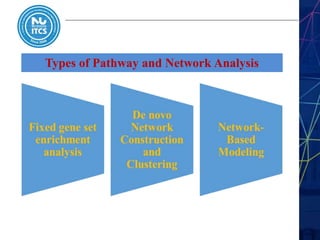
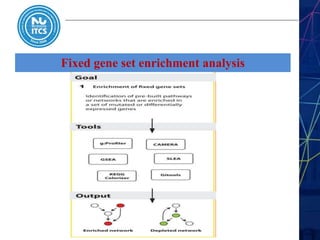
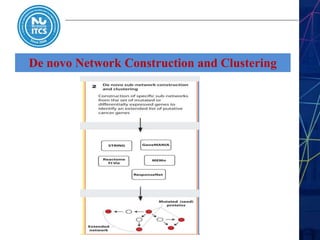
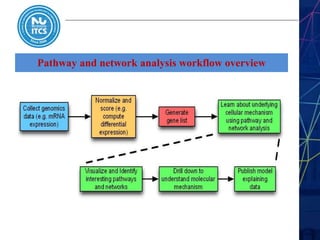
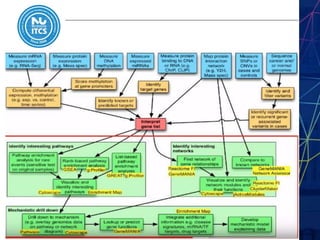
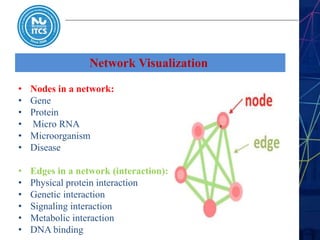
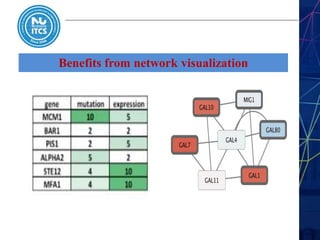
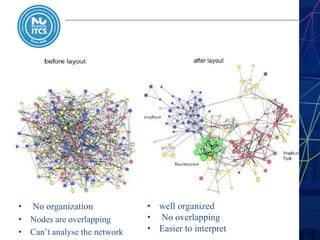
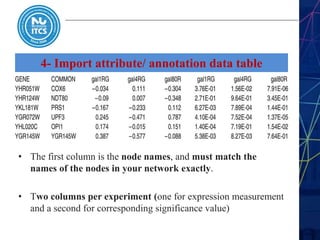
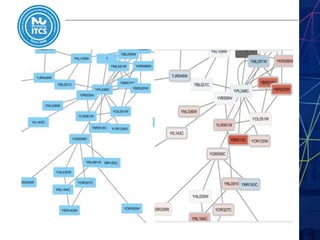
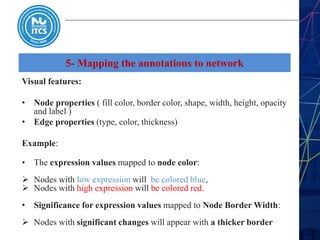
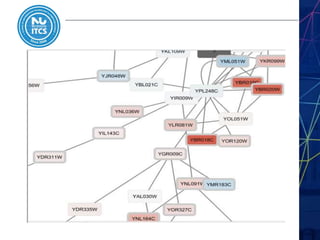
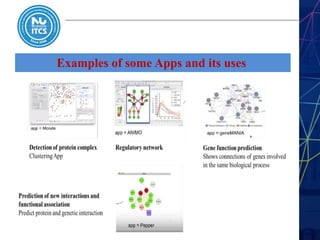
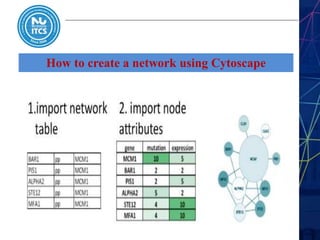
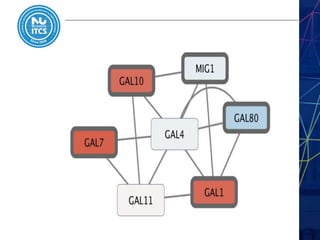
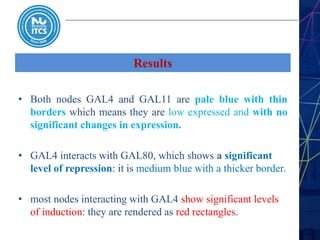
This document discusses pathway and network analysis. It defines systems biology and biological networks. Some benefits of studying pathways and networks are that it improves statistical power, allows identification of potential causal mechanisms, and facilitates integration of multiple data types. Types of analysis include gene set enrichment and de novo network construction. Visualization is important for representing relationships between molecules and finding subnetworks. Software like Cytoscape can be used to import networks, map gene expression data to node colors/borders, filter networks, and export publication-quality images. A tutorial demonstrates combining expression and network data in Cytoscape to tell biological stories.